fitrkernel
Fit Gaussian kernel regression model using random feature expansion
Syntax
Description
fitrkernel trains or cross-validates a Gaussian kernel
regression model for nonlinear regression. fitrkernel is more
practical to use for big data applications that have large training sets, but can also
be applied to smaller data sets that fit in memory.
fitrkernel maps data in a low-dimensional space into a
high-dimensional space, then fits a linear model in the high-dimensional space by
minimizing the regularized objective function. Obtaining the linear model in the
high-dimensional space is equivalent to applying the Gaussian kernel to the model in the
low-dimensional space. Available linear regression models include regularized support
vector machine (SVM) and least-squares regression models.
To train a nonlinear SVM regression model on in-memory data, see fitrsvm.
Mdl = fitrkernel(Tbl,ResponseVarName)Mdl trained using the
predictor variables contained in the table Tbl and the
response values in Tbl.ResponseVarName.
Mdl = fitrkernel(___,Name,Value)
[
also returns the hyperparameter optimization results when you specify
Mdl,FitInfo,HyperparameterOptimizationResults] = fitrkernel(___)OptimizeHyperparameters.
[
also returns Mdl,FitInfo,AggregateOptimizationResults] = fitrkernel(___)AggregateOptimizationResults, which contains
hyperparameter optimization results when you specify the
OptimizeHyperparameters and
HyperparameterOptimizationOptions name-value arguments.
You must also specify the ConstraintType and
ConstraintBounds options of
HyperparameterOptimizationOptions. You can use this
syntax to optimize on compact model size instead of cross-validation loss, and
to perform a set of multiple optimization problems that have the same options
but different constraint bounds.
Examples
Train a kernel regression model for a tall array by using SVM.
When you perform calculations on tall arrays, MATLAB® uses either a parallel pool (default if you have Parallel Computing Toolbox™) or the local MATLAB session. To run the example using the local MATLAB session when you have Parallel Computing Toolbox, change the global execution environment by using the mapreducer function.
mapreducer(0)
Create a datastore that references the folder location with the data. The data can be contained in a single file, a collection of files, or an entire folder. Treat 'NA' values as missing data so that datastore replaces them with NaN values. Select a subset of the variables to use. Create a tall table on top of the datastore.
varnames = {'ArrTime','DepTime','ActualElapsedTime'};
ds = datastore('airlinesmall.csv','TreatAsMissing','NA',...
'SelectedVariableNames',varnames);
t = tall(ds);Specify DepTime and ArrTime as the predictor variables (X) and ActualElapsedTime as the response variable (Y). Select the observations for which ArrTime is later than DepTime.
daytime = t.ArrTime>t.DepTime; Y = t.ActualElapsedTime(daytime); % Response data X = t{daytime,{'DepTime' 'ArrTime'}}; % Predictor data
Standardize the predictor variables.
Z = zscore(X); % Standardize the dataTrain a default Gaussian kernel regression model with the standardized predictors. Extract a fit summary to determine how well the optimization algorithm fits the model to the data.
[Mdl,FitInfo] = fitrkernel(Z,Y)
Found 6 chunks. |========================================================================= | Solver | Iteration / | Objective | Gradient | Beta relative | | | Data Pass | | magnitude | change | |========================================================================= | INIT | 0 / 1 | 4.307833e+01 | 9.925486e-02 | NaN | | LBFGS | 0 / 2 | 2.782790e+01 | 7.202403e-03 | 9.891473e-01 | | LBFGS | 1 / 3 | 2.781351e+01 | 1.806211e-02 | 3.220672e-03 | | LBFGS | 2 / 4 | 2.777773e+01 | 2.727737e-02 | 9.309939e-03 | | LBFGS | 3 / 5 | 2.768591e+01 | 2.951422e-02 | 2.833343e-02 | | LBFGS | 4 / 6 | 2.755857e+01 | 5.124144e-02 | 7.935278e-02 | | LBFGS | 5 / 7 | 2.738896e+01 | 3.089571e-02 | 4.644920e-02 | | LBFGS | 6 / 8 | 2.716704e+01 | 2.552696e-02 | 8.596406e-02 | | LBFGS | 7 / 9 | 2.696409e+01 | 3.088621e-02 | 1.263589e-01 | | LBFGS | 8 / 10 | 2.676203e+01 | 2.021303e-02 | 1.533927e-01 | | LBFGS | 9 / 11 | 2.660322e+01 | 1.221361e-02 | 1.351968e-01 | | LBFGS | 10 / 12 | 2.645504e+01 | 1.486501e-02 | 1.175476e-01 | | LBFGS | 11 / 13 | 2.631323e+01 | 1.772835e-02 | 1.161909e-01 | | LBFGS | 12 / 14 | 2.625264e+01 | 5.837906e-02 | 1.422851e-01 | | LBFGS | 13 / 15 | 2.619281e+01 | 1.294441e-02 | 2.966283e-02 | | LBFGS | 14 / 16 | 2.618220e+01 | 3.791806e-03 | 9.051274e-03 | | LBFGS | 15 / 17 | 2.617989e+01 | 3.689255e-03 | 6.364132e-03 | | LBFGS | 16 / 18 | 2.617426e+01 | 4.200232e-03 | 1.213026e-02 | | LBFGS | 17 / 19 | 2.615914e+01 | 7.339928e-03 | 2.803348e-02 | | LBFGS | 18 / 20 | 2.620704e+01 | 2.298098e-02 | 1.749830e-01 | |========================================================================= | Solver | Iteration / | Objective | Gradient | Beta relative | | | Data Pass | | magnitude | change | |========================================================================= | LBFGS | 18 / 21 | 2.615554e+01 | 1.164689e-02 | 8.580878e-02 | | LBFGS | 19 / 22 | 2.614367e+01 | 3.395507e-03 | 3.938314e-02 | | LBFGS | 20 / 23 | 2.614090e+01 | 2.349246e-03 | 1.495049e-02 | |========================================================================|
Mdl =
RegressionKernel
ResponseName: 'Y'
Learner: 'svm'
NumExpansionDimensions: 64
KernelScale: 1
Lambda: 8.5385e-06
BoxConstraint: 1
Epsilon: 5.9303
Properties, Methods
FitInfo = struct with fields:
Solver: 'LBFGS-tall'
LossFunction: 'epsiloninsensitive'
Lambda: 8.5385e-06
BetaTolerance: 1.0000e-03
GradientTolerance: 1.0000e-05
ObjectiveValue: 26.1409
GradientMagnitude: 0.0023
RelativeChangeInBeta: 0.0150
FitTime: 9.1186
History: [1×1 struct]
Mdl is a RegressionKernel model. To inspect the regression error, you can pass Mdl and the training data or new data to the loss function. Or, you can pass Mdl and new predictor data to the predict function to predict responses for new observations. You can also pass Mdl and the training data to the resume function to continue training.
FitInfo is a structure array containing optimization information. Use FitInfo to determine whether optimization termination measurements are satisfactory.
For improved accuracy, you can increase the maximum number of optimization iterations ('IterationLimit') and decrease the tolerance values ('BetaTolerance' and 'GradientTolerance') by using the name-value pair arguments of fitrkernel. Doing so can improve measures like ObjectiveValue and RelativeChangeInBeta in FitInfo. You can also optimize model parameters by using the 'OptimizeHyperparameters' name-value pair argument.
Load the carbig data set.
load carbigSpecify the predictor variables (X) and the response variable (Y).
X = [Acceleration,Cylinders,Displacement,Horsepower,Weight]; Y = MPG;
Delete rows of X and Y where either array has NaN values. Removing rows with NaN values before passing data to fitrkernel can speed up training and reduce memory usage.
R = rmmissing([X Y]); % Data with missing entries removed
X = R(:,1:5);
Y = R(:,end); Cross-validate a kernel regression model using 5-fold cross-validation. Standardize the predictor variables.
Mdl = fitrkernel(X,Y,'Kfold',5,'Standardize',true)
Mdl =
RegressionPartitionedKernel
CrossValidatedModel: 'Kernel'
ResponseName: 'Y'
NumObservations: 392
KFold: 5
Partition: [1×1 cvpartition]
ResponseTransform: 'none'
Properties, Methods
numel(Mdl.Trained)
ans = 5
Mdl is a RegressionPartitionedKernel model. Because fitrkernel implements five-fold cross-validation, Mdl contains five RegressionKernel models that the software trains on training-fold (in-fold) observations.
Examine the cross-validation loss (mean squared error) for each fold.
kfoldLoss(Mdl,'mode','individual')
ans = 5×1
13.1983
14.2686
23.9162
21.0763
24.3975
Optimize hyperparameters automatically using the OptimizeHyperparameters name-value argument.
Load the carbig data set.
load carbigSpecify the predictor variables (X) and the response variable (Y).
X = [Acceleration,Cylinders,Displacement,Horsepower,Weight]; Y = MPG;
Delete rows of X and Y where either array has NaN values. Removing rows with NaN values before passing data to fitrkernel can speed up training and reduce memory usage.
R = rmmissing([X Y]); % Data with missing entries removed
X = R(:,1:5);
Y = R(:,end); Find hyperparameters that minimize five-fold cross-validation loss by using automatic hyperparameter optimization. Specify OptimizeHyperparameters as 'auto' so that fitrkernel finds the optimal values of the KernelScale, Lambda, Epsilon, and Standardize name-value arguments. For reproducibility, set the random seed and use the 'expected-improvement-plus' acquisition function.
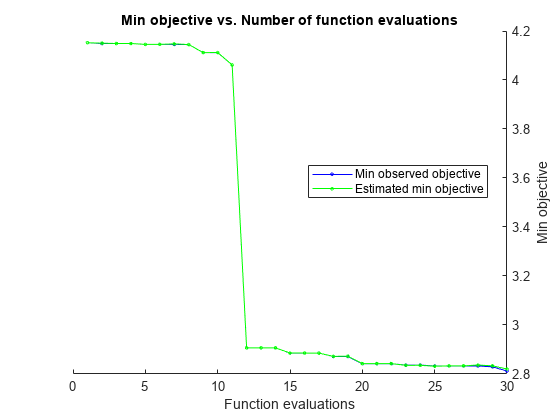
rng('default') [Mdl,FitInfo,HyperparameterOptimizationResults] = fitrkernel(X,Y,'OptimizeHyperparameters','auto',... 'HyperparameterOptimizationOptions',struct('AcquisitionFunctionName','expected-improvement-plus'))
|===================================================================================================================================|
| Iter | Eval | Objective: | Objective | BestSoFar | BestSoFar | KernelScale | Lambda | Epsilon | Standardize |
| | result | log(1+loss) | runtime | (observed) | (estim.) | | | | |
|===================================================================================================================================|
| 1 | Best | 4.1521 | 0.32486 | 4.1521 | 4.1521 | 11.415 | 0.0017304 | 615.77 | true |
| 2 | Best | 4.1489 | 0.095056 | 4.1489 | 4.1503 | 509.07 | 0.0064454 | 0.048411 | true |
| 3 | Accept | 5.251 | 0.58842 | 4.1489 | 4.1489 | 0.0015621 | 1.8257e-05 | 0.051954 | true |
| 4 | Accept | 4.3329 | 0.11052 | 4.1489 | 4.1489 | 0.0053278 | 2.37 | 17.883 | false |
| 5 | Accept | 4.2414 | 0.11175 | 4.1489 | 4.1489 | 0.004474 | 0.13531 | 14.426 | true |
| 6 | Best | 4.148 | 0.11424 | 4.148 | 4.148 | 0.43562 | 2.5339 | 0.059928 | true |
| 7 | Accept | 4.1521 | 0.083724 | 4.148 | 4.148 | 3.2193 | 0.012683 | 813.56 | false |
| 8 | Best | 3.8438 | 0.095093 | 3.8438 | 3.8439 | 5.7821 | 0.065897 | 2.056 | true |
| 9 | Accept | 4.1305 | 0.12469 | 3.8438 | 3.8439 | 110.96 | 0.42454 | 7.6606 | true |
| 10 | Best | 3.7951 | 0.10216 | 3.7951 | 3.7954 | 1.1595 | 0.054292 | 0.012493 | true |
| 11 | Accept | 4.2311 | 0.39632 | 3.7951 | 3.7954 | 0.0011423 | 0.00015862 | 8.6125 | false |
| 12 | Best | 2.8871 | 0.80112 | 2.8871 | 2.8872 | 185.22 | 2.1981e-05 | 1.0401 | false |
| 13 | Accept | 4.1521 | 0.080121 | 2.8871 | 3.0058 | 993.92 | 2.6036e-06 | 58.773 | false |
| 14 | Best | 2.8648 | 0.6215 | 2.8648 | 2.8765 | 196.57 | 2.2026e-05 | 1.081 | false |
| 15 | Accept | 4.2977 | 0.1799 | 2.8648 | 2.8668 | 0.017949 | 1.5685e-05 | 15.01 | false |
| 16 | Best | 2.8016 | 0.46806 | 2.8016 | 2.8017 | 786 | 3.4462e-06 | 1.6117 | false |
| 17 | Accept | 2.9032 | 0.23372 | 2.8016 | 2.8026 | 974.16 | 0.00019486 | 1.6661 | false |
| 18 | Accept | 2.9051 | 0.95934 | 2.8016 | 2.8018 | 288.21 | 2.6218e-06 | 2.0933 | false |
| 19 | Accept | 3.4438 | 1.1165 | 2.8016 | 2.803 | 56.999 | 2.885e-06 | 1.3903 | false |
| 20 | Accept | 2.8436 | 0.91882 | 2.8016 | 2.8032 | 533.99 | 2.7293e-06 | 0.6719 | false |
|===================================================================================================================================|
| Iter | Eval | Objective: | Objective | BestSoFar | BestSoFar | KernelScale | Lambda | Epsilon | Standardize |
| | result | log(1+loss) | runtime | (observed) | (estim.) | | | | |
|===================================================================================================================================|
| 21 | Accept | 2.8301 | 0.65452 | 2.8016 | 2.8024 | 411.02 | 3.4347e-06 | 0.98949 | false |
| 22 | Accept | 2.8233 | 0.33307 | 2.8016 | 2.8043 | 455.25 | 5.2936e-05 | 1.1189 | false |
| 23 | Accept | 4.1168 | 0.12289 | 2.8016 | 2.802 | 237.02 | 0.85493 | 0.42894 | false |
| 24 | Best | 2.7876 | 0.49534 | 2.7876 | 2.7877 | 495.51 | 1.8049e-05 | 1.9006 | false |
| 25 | Accept | 2.8197 | 0.31334 | 2.7876 | 2.7877 | 927.29 | 1.128e-05 | 1.1902 | false |
| 26 | Accept | 2.8361 | 0.32645 | 2.7876 | 2.7882 | 354.44 | 6.1939e-05 | 2.2591 | false |
| 27 | Accept | 2.7985 | 0.67968 | 2.7876 | 2.7906 | 506.54 | 1.4142e-05 | 1.3659 | false |
| 28 | Accept | 2.8163 | 0.38706 | 2.7876 | 2.7905 | 829.6 | 1.0965e-05 | 2.7415 | false |
| 29 | Accept | 2.8469 | 0.57559 | 2.7876 | 2.7902 | 729.48 | 3.4914e-06 | 0.039087 | false |
| 30 | Accept | 2.882 | 0.82757 | 2.7876 | 2.7902 | 255.25 | 3.2869e-06 | 0.059794 | false |
__________________________________________________________
Optimization completed.
MaxObjectiveEvaluations of 30 reached.
Total function evaluations: 30
Total elapsed time: 21.3944 seconds
Total objective function evaluation time: 12.2415
Best observed feasible point:
KernelScale Lambda Epsilon Standardize
___________ __________ _______ ___________
495.51 1.8049e-05 1.9006 false
Observed objective function value = 2.7876
Estimated objective function value = 2.7902
Function evaluation time = 0.49534
Best estimated feasible point (according to models):
KernelScale Lambda Epsilon Standardize
___________ __________ _______ ___________
495.51 1.8049e-05 1.9006 false
Estimated objective function value = 2.7902
Estimated function evaluation time = 0.45863
Mdl =
RegressionKernel
ResponseName: 'Y'
Learner: 'svm'
NumExpansionDimensions: 256
KernelScale: 495.5140
Lambda: 1.8049e-05
BoxConstraint: 141.3376
Epsilon: 1.9006
Properties, Methods
FitInfo = struct with fields:
Solver: 'LBFGS-fast'
LossFunction: 'epsiloninsensitive'
Lambda: 1.8049e-05
BetaTolerance: 1.0000e-04
GradientTolerance: 1.0000e-06
ObjectiveValue: 1.3382
GradientMagnitude: 0.0051
RelativeChangeInBeta: 9.4332e-05
FitTime: 0.0462
History: []
HyperparameterOptimizationResults =
BayesianOptimization with properties:
ObjectiveFcn: @createObjFcn/inMemoryObjFcn
VariableDescriptions: [6×1 optimizableVariable]
Options: [1×1 struct]
MinObjective: 2.7876
XAtMinObjective: [1×4 table]
MinEstimatedObjective: 2.7902
XAtMinEstimatedObjective: [1×4 table]
NumObjectiveEvaluations: 30
TotalElapsedTime: 21.3944
NextPoint: [1×4 table]
XTrace: [30×4 table]
ObjectiveTrace: [30×1 double]
ConstraintsTrace: []
UserDataTrace: {30×1 cell}
ObjectiveEvaluationTimeTrace: [30×1 double]
IterationTimeTrace: [30×1 double]
ErrorTrace: [30×1 double]
FeasibilityTrace: [30×1 logical]
FeasibilityProbabilityTrace: [30×1 double]
IndexOfMinimumTrace: [30×1 double]
ObjectiveMinimumTrace: [30×1 double]
EstimatedObjectiveMinimumTrace: [30×1 double]
For big data, the optimization procedure can take a long time. If the data set is too large to run the optimization procedure, you can try to optimize the parameters using only partial data. Use the datasample function and specify 'Replace','false' to sample data without replacement.
Input Arguments
Predictor data to which the regression model is fit, specified as an n-by-p numeric matrix, where n is the number of observations and p is the number of predictor variables.
The length of Y and the number of observations in
X must be equal.
Data Types: single | double
Sample data used to train the model, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor variable.
Optionally, Tbl can contain one additional column for the response
variable. Multicolumn variables and cell arrays other than cell arrays of character
vectors are not allowed.
If
Tblcontains the response variable, and you want to use all remaining variables inTblas predictors, then specify the response variable by usingResponseVarName.If
Tblcontains the response variable, and you want to use only a subset of the remaining variables inTblas predictors, then specify a formula by usingformula.If
Tbldoes not contain the response variable, then specify a response variable by usingY. The length of the response variable and the number of rows inTblmust be equal.
Response variable name, specified as the name of a variable in
Tbl. The response variable must be a numeric vector.
You must specify ResponseVarName as a character vector or string
scalar. For example, if Tbl stores the response variable
Y as Tbl.Y, then specify it as
"Y". Otherwise, the software treats all columns of
Tbl, including Y, as predictors when
training the model.
Data Types: char | string
Explanatory model of the response variable and a subset of the predictor variables,
specified as a character vector or string scalar in the form
"Y~x1+x2+x3". In this form, Y represents the
response variable, and x1, x2, and
x3 represent the predictor variables.
To specify a subset of variables in Tbl as predictors for
training the model, use a formula. If you specify a formula, then the software does not
use any variables in Tbl that do not appear in
formula.
The variable names in the formula must be both variable names in Tbl
(Tbl.Properties.VariableNames) and valid MATLAB® identifiers. You can verify the variable names in Tbl by
using the isvarname function. If the variable names
are not valid, then you can convert them by using the matlab.lang.makeValidName function.
Data Types: char | string
Note
The software treats NaN, empty character vector
(''), empty string (""),
<missing>, and <undefined>
elements as missing values, and removes observations with any of these characteristics:
Missing value in the response variable
At least one missing value in a predictor observation (row in
XorTbl)NaNvalue or0weight ('Weights')
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: Mdl =
fitrkernel(X,Y,Learner="leastsquares",NumExpansionDimensions=2^15,KernelScale="auto")
implements least-squares regression after mapping the predictor data to the
2^15 dimensional space using feature expansion with a kernel
scale parameter selected by a heuristic procedure.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: Mdl =
fitrkernel(X,Y,'Learner','leastsquares','NumExpansionDimensions',2^15,'KernelScale','auto')
Note
You cannot use any cross-validation name-value argument together with the
OptimizeHyperparameters name-value argument. You can modify the
cross-validation for OptimizeHyperparameters only by using the
HyperparameterOptimizationOptions name-value argument.
Kernel Regression Options
Box
constraint, specified as the comma-separated pair consisting
of 'BoxConstraint' and a positive scalar.
This argument is valid only when 'Learner' is
'svm'(default) and you do not specify a value for
the regularization term strength 'Lambda'. You can
specify either 'BoxConstraint' or
'Lambda' because the box constraint
(C) and the regularization term strength
(λ) are related by C =
1/(λn), where n is the number of
observations (rows in X).
Example: 'BoxConstraint',100
Data Types: single | double
Half the width of the epsilon-insensitive band, specified as the
comma-separated pair consisting of 'Epsilon' and
'auto' or a nonnegative scalar value.
For 'auto', the fitrkernel
function determines the value of Epsilon as
iqr(Y)/13.49, which is an estimate of a tenth of
the standard deviation using the interquartile range of the response
variable Y. If iqr(Y) is equal to
zero, then fitrkernel sets the value of
Epsilon to 0.1.
'Epsilon' is valid only when
Learner is svm.
Example: 'Epsilon',0.3
Data Types: single | double
Number of dimensions of the expanded space, specified as the
comma-separated pair consisting of
'NumExpansionDimensions' and
'auto' or a positive integer. For
'auto', the fitrkernel
function selects the number of dimensions using
2.^ceil(min(log2(p)+5,15)), where
p is the number of predictors.
Example: 'NumExpansionDimensions',2^15
Data Types: char | string | single | double
Kernel scale parameter, specified as the comma-separated pair
consisting of 'KernelScale' and
'auto' or a positive scalar. MATLAB obtains the random basis for random feature expansion by
using the kernel scale parameter. For details, see Random Feature Expansion.
If you specify 'auto', then MATLAB selects an appropriate kernel scale parameter using a
heuristic procedure. This heuristic procedure uses subsampling, so
estimates can vary from one call to another. Therefore, to reproduce
results, set a random number seed by using rng before
training.
Example: 'KernelScale','auto'
Data Types: char | string | single | double
Regularization term strength, specified as the comma-separated pair consisting of 'Lambda' and 'auto' or a nonnegative scalar.
For 'auto', the value of Lambda is
1/n, where n is the number of
observations.
When Learner is 'svm', you can specify either
BoxConstraint or Lambda because the box
constraint (C) and the regularization term strength
(λ) are related by C =
1/(λn).
Example: 'Lambda',0.01
Data Types: char | string | single | double
Linear regression model type, specified as the comma-separated pair
consisting of 'Learner' and 'svm'
or 'leastsquares'.
In the following table,
x is an observation (row vector) from p predictor variables.
is a transformation of an observation (row vector) for feature expansion. T(x) maps x in to a high-dimensional space ().
β is a vector of coefficients.
b is the scalar bias.
| Value | Algorithm | Response range | Loss function |
|---|---|---|---|
'leastsquares' | Linear regression via ordinary least squares | y ∊ (-∞,∞) | Mean squared error (MSE): |
'svm' | Support vector machine regression | Same as 'leastsquares' | Epsilon-insensitive: |
Example: 'Learner','leastsquares'
Since R2023b
Flag to standardize the predictor data, specified as a numeric or logical 0
(false) or 1 (true). If you
set Standardize to true, then the software
centers and scales each numeric predictor variable by the corresponding column mean and
standard deviation. The software does not standardize the categorical predictors.
Example: "Standardize",true
Data Types: single | double | logical
Verbosity level, specified as the comma-separated pair consisting of
'Verbose' and either 0 or
1. Verbose controls the amount
of diagnostic information fitrkernel displays at
the command line.
| Value | Description |
|---|---|
0 | fitrkernel does not display
diagnostic information. |
1 | fitrkernel displays and stores
the value of the objective function, gradient magnitude,
and other diagnostic information.
FitInfo.History contains the
diagnostic information. |
Example: 'Verbose',1
Data Types: single | double
Maximum amount of allocated memory (in megabytes), specified as the
comma-separated pair consisting of 'BlockSize' and a
positive scalar.
If fitrkernel requires more memory than the value
of BlockSize to hold the transformed predictor data,
then MATLAB uses a block-wise strategy. For details about the
block-wise strategy, see Algorithms.
Example: 'BlockSize',1e4
Data Types: single | double
Random number stream for reproducibility of data transformation,
specified as the comma-separated pair consisting of
'RandomStream' and a random stream object. For
details, see Random Feature Expansion.
Use 'RandomStream' to reproduce the random basis
functions that fitrkernel uses to transform the data
in X to a high-dimensional space. For details, see
Managing the Global Stream Using RandStream and Creating and Controlling a Random Number Stream.
Example: 'RandomStream',RandStream('mlfg6331_64')
Other Regression Options
Categorical predictors list, specified as one of the values in this table.
| Value | Description |
|---|---|
| Vector of positive integers |
Each entry in the vector is an index value indicating that the corresponding predictor is
categorical. The index values are between 1 and If |
| Logical vector |
A |
| Character matrix | Each row of the matrix is the name of a predictor variable. The names must match the entries in PredictorNames. Pad the names with extra blanks so each row of the character matrix has the same length. |
| String array or cell array of character vectors | Each element in the array is the name of a predictor variable. The names must match the entries in PredictorNames. |
"all" | All predictors are categorical. |
By default, if the
predictor data is in a table (Tbl), fitrkernel
assumes that a variable is categorical if it is a logical vector, categorical vector, character
array, string array, or cell array of character vectors. If the predictor data is a matrix
(X), fitrkernel assumes that all predictors are
continuous. To identify any other predictors as categorical predictors, specify them by using
the CategoricalPredictors name-value argument.
For the identified categorical predictors, fitrkernel creates dummy variables using two different schemes, depending on whether a categorical variable is unordered or ordered. For an unordered categorical variable, fitrkernel creates one dummy variable for each level of the categorical variable. For an ordered categorical variable, fitrkernel creates one less dummy variable than the number of categories. For details, see Automatic Creation of Dummy Variables.
Example: 'CategoricalPredictors','all'
Data Types: single | double | logical | char | string | cell
Predictor variable names, specified as a string array of unique names or cell array of unique
character vectors. The functionality of PredictorNames depends on the
way you supply the training data.
If you supply
XandY, then you can usePredictorNamesto assign names to the predictor variables inX.The order of the names in
PredictorNamesmust correspond to the column order ofX. That is,PredictorNames{1}is the name ofX(:,1),PredictorNames{2}is the name ofX(:,2), and so on. Also,size(X,2)andnumel(PredictorNames)must be equal.By default,
PredictorNamesis{'x1','x2',...}.
If you supply
Tbl, then you can usePredictorNamesto choose which predictor variables to use in training. That is,fitrkerneluses only the predictor variables inPredictorNamesand the response variable during training.PredictorNamesmust be a subset ofTbl.Properties.VariableNamesand cannot include the name of the response variable.By default,
PredictorNamescontains the names of all predictor variables.A good practice is to specify the predictors for training using either
PredictorNamesorformula, but not both.
Example: "PredictorNames",["SepalLength","SepalWidth","PetalLength","PetalWidth"]
Data Types: string | cell
Response variable name, specified as a character vector or string scalar.
If you supply
Y, then you can useResponseNameto specify a name for the response variable.If you supply
ResponseVarNameorformula, then you cannot useResponseName.
Example: ResponseName="response"
Data Types: char | string
Function for transforming raw response values, specified as a function handle or
function name. The default is "none", which means
@(y)y, or no transformation. The function should accept a vector
(the original response values) and return a vector of the same size (the transformed
response values).
Example: Suppose you create a function handle that applies an exponential
transformation to an input vector by using myfunction = @(y)exp(y).
Then, you can specify the response transformation as
ResponseTransform=myfunction.
Data Types: char | string | function_handle
Observation weights, specified as the comma-separated pair consisting
of 'Weights' and a vector of scalar values or the
name of a variable in Tbl. The software weights
each observation (or row) in X or
Tbl with the corresponding value in
Weights. The length of
Weights must equal the number of rows in
X or Tbl.
If you specify the input data as a table Tbl,
then Weights can be the name of a variable in
Tbl that contains a numeric vector. In this
case, you must specify Weights as a character
vector or string scalar. For example, if weights vector
W is stored as Tbl.W, then
specify it as 'W'. Otherwise, the software treats all
columns of Tbl, including W, as
predictors when training the model.
By default, Weights is
ones(n,1), where n is the
number of observations in X or
Tbl.
fitrkernel normalizes the weights to sum to 1.
Inf weights are not supported.
Data Types: single | double | char | string
Cross-Validation Options
Cross-validation flag, specified as the comma-separated pair
consisting of 'Crossval' and 'on'
or 'off'.
If you specify 'on', then the software implements
10-fold cross-validation.
You can override this cross-validation setting using the
CVPartition, Holdout,
KFold, or Leaveout
name-value pair argument. You can use only one cross-validation
name-value pair argument at a time to create a cross-validated
model.
Example: 'Crossval','on'
Cross-validation partition, specified as a cvpartition object that specifies the type of cross-validation and the
indexing for the training and validation sets.
To create a cross-validated model, you can specify only one of these four name-value
arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: Suppose you create a random partition for 5-fold cross-validation on 500
observations by using cvp = cvpartition(500,KFold=5). Then, you can
specify the cross-validation partition by setting
CVPartition=cvp.
Fraction of the data used for holdout validation, specified as a scalar value in the range
(0,1). If you specify Holdout=p, then the software completes these
steps:
Randomly select and reserve
p*100% of the data as validation data, and train the model using the rest of the data.Store the compact trained model in the
Trainedproperty of the cross-validated model.
To create a cross-validated model, you can specify only one of these four name-value
arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: Holdout=0.1
Data Types: double | single
Number of folds to use in the cross-validated model, specified as a positive integer value
greater than 1. If you specify KFold=k, then the software completes
these steps:
Randomly partition the data into
ksets.For each set, reserve the set as validation data, and train the model using the other
k– 1 sets.Store the
kcompact trained models in ak-by-1 cell vector in theTrainedproperty of the cross-validated model.
To create a cross-validated model, you can specify only one of these four name-value
arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: KFold=5
Data Types: single | double
Leave-one-out cross-validation flag, specified as the comma-separated pair consisting of
'Leaveout' and 'on' or
'off'. If you specify 'Leaveout','on', then,
for each of the n observations (where n is the
number of observations excluding missing observations), the software completes these
steps:
Reserve the observation as validation data, and train the model using the other n – 1 observations.
Store the n compact, trained models in the cells of an n-by-1 cell vector in the
Trainedproperty of the cross-validated model.
To create a cross-validated model, you can use one of these
four name-value pair arguments only: CVPartition, Holdout, KFold,
or Leaveout.
Example: 'Leaveout','on'
Convergence Controls
Relative tolerance on the linear coefficients and the bias term (intercept), specified as a nonnegative scalar.
Let , that is, the vector of the coefficients and the bias term at optimization iteration t. If , then optimization terminates.
If you also specify GradientTolerance, then optimization terminates when the software satisfies either stopping criterion.
Example: 'BetaTolerance',1e-6
Data Types: single | double
Absolute gradient tolerance, specified as a nonnegative scalar.
Let be the gradient vector of the objective function with respect to the coefficients and bias term at optimization iteration t. If , then optimization terminates.
If you also specify BetaTolerance, then optimization terminates when the
software satisfies either stopping criterion.
Example: 'GradientTolerance',1e-5
Data Types: single | double
Size of the history buffer for Hessian approximation, specified as the
comma-separated pair consisting of
'HessianHistorySize' and a positive integer. At
each iteration, fitrkernel composes the Hessian by
using statistics from the latest HessianHistorySize
iterations.
Example: 'HessianHistorySize',10
Data Types: single | double
Maximum number of optimization iterations, specified as the
comma-separated pair consisting of 'IterationLimit'
and a positive integer.
The default value is 1000 if the transformed data fits in memory, as
specified by BlockSize. Otherwise, the default
value is 100.
Example: 'IterationLimit',500
Data Types: single | double
Hyperparameter Optimization Options
Parameters to optimize, specified as the comma-separated pair
consisting of 'OptimizeHyperparameters' and one of
these values:
'none'— Do not optimize.'auto'— Use{'KernelScale','Lambda','Epsilon','Standardize'}.'all'— Optimize all eligible parameters.Cell array of eligible parameter names.
Vector of
optimizableVariableobjects, typically the output ofhyperparameters.
The optimization attempts to minimize the cross-validation loss
(error) for fitrkernel by varying the parameters. To control the
cross-validation type and other aspects of the optimization, use the
HyperparameterOptimizationOptions name-value argument. When you use
HyperparameterOptimizationOptions, you can use the (compact) model size
instead of the cross-validation loss as the optimization objective by setting the
ConstraintType and ConstraintBounds options.
Note
The values of OptimizeHyperparameters override any values you
specify using other name-value arguments. For example, setting
OptimizeHyperparameters to "auto" causes
fitrkernel to optimize hyperparameters corresponding to the
"auto" option and to ignore any specified values for the
hyperparameters.
The eligible parameters for fitrkernel
are:
Epsilon—fitrkernelsearches among positive values, by default log-scaled in the range[1e-3,1e2]*iqr(Y)/1.349.KernelScale—fitrkernelsearches among positive values, by default log-scaled in the range[1e-3,1e3].Lambda—fitrkernelsearches among positive values, by default log-scaled in the range[1e-3,1e3]/n, wherenis the number of observations.Learner—fitrkernelsearches among'svm'and'leastsquares'.NumExpansionDimensions—fitrkernelsearches among positive integers, by default log-scaled in the range[100,10000].Standardize—fitrkernelsearches amongtrueandfalse.
Set nondefault parameters by passing a vector of
optimizableVariable objects that have nondefault
values. For example:
load carsmall params = hyperparameters('fitrkernel',[Horsepower,Weight],MPG); params(2).Range = [1e-4,1e6];
Pass params as the value of
'OptimizeHyperparameters'.
By default, the iterative display appears at the command line,
and plots appear according to the number of hyperparameters in the optimization. For the
optimization and plots, the objective function is log(1 + cross-validation loss). To control the iterative display, set the Verbose option
of the HyperparameterOptimizationOptions name-value argument. To control
the plots, set the ShowPlots option of the
HyperparameterOptimizationOptions name-value argument.
For an example, see Optimize Kernel Regression.
Example: 'OptimizeHyperparameters','auto'
Options for optimization, specified as a HyperparameterOptimizationOptions object or a structure. This argument
modifies the effect of the OptimizeHyperparameters name-value
argument. If you specify HyperparameterOptimizationOptions, you must
also specify OptimizeHyperparameters. All the options are optional.
However, you must set ConstraintBounds and
ConstraintType to return
AggregateOptimizationResults. The options that you can set in a
structure are the same as those in the
HyperparameterOptimizationOptions object.
| Option | Values | Default |
|---|---|---|
Optimizer |
| "bayesopt" |
ConstraintBounds | Constraint bounds for N optimization problems,
specified as an N-by-2 numeric matrix or
| [] |
ConstraintTarget | Constraint target for the optimization problems, specified as
| If you specify ConstraintBounds and
ConstraintType, then the default value is
"matlab". Otherwise, the default value is
[]. |
ConstraintType | Constraint type for the optimization problems, specified as
| [] |
AcquisitionFunctionName | Type of acquisition function:
Acquisition functions whose names include
| "expected-improvement-per-second-plus" |
MaxObjectiveEvaluations | Maximum number of objective function evaluations. If you specify multiple
optimization problems using ConstraintBounds, the value of
MaxObjectiveEvaluations applies to each optimization
problem individually. | 30 for "bayesopt" and
"randomsearch", and the entire grid for
"gridsearch" |
MaxTime | Time limit for the optimization, specified as a nonnegative real
scalar. The time limit is in seconds, as measured by | Inf |
NumGridDivisions | For Optimizer="gridsearch", the number of values in each
dimension. The value can be a vector of positive integers giving the number of
values for each dimension, or a scalar that applies to all dimensions. The
software ignores this option for categorical variables. | 10 |
ShowPlots | Logical value indicating whether to show plots of the optimization progress.
If this option is true, the software plots the best observed
objective function value against the iteration number. If you use Bayesian
optimization (Optimizer="bayesopt"), the
software also plots the best estimated objective function value. The best
observed objective function values and best estimated objective function values
correspond to the values in the BestSoFar (observed) and
BestSoFar (estim.) columns of the iterative display,
respectively. You can find these values in the properties ObjectiveMinimumTrace and EstimatedObjectiveMinimumTrace of
Mdl.HyperparameterOptimizationResults. If the problem
includes one or two optimization parameters for Bayesian optimization, then
ShowPlots also plots a model of the objective function
against the parameters. | true |
SaveIntermediateResults | Logical value indicating whether to save the optimization results. If this
option is true, the software overwrites a workspace variable
named "BayesoptResults" at each iteration. The variable is a
BayesianOptimization object. If you
specify multiple optimization problems using
ConstraintBounds, the workspace variable is an AggregateBayesianOptimization object named
"AggregateBayesoptResults". | false |
Verbose | Display level at the command line:
For details, see the | 1 |
UseParallel | Logical value indicating whether to run the Bayesian optimization in parallel, which requires Parallel Computing Toolbox™. Due to the nonreproducibility of parallel timing, parallel Bayesian optimization does not necessarily yield reproducible results. For details, see Parallel Bayesian Optimization. | false |
Repartition | Logical value indicating whether to repartition the cross-validation at
every iteration. If this option is A value of
| false |
| Specify only one of the following three options. | ||
CVPartition | cvpartition object created by cvpartition | KFold=5 if you do not specify a
cross-validation option |
Holdout | Scalar in the range (0,1) representing the holdout
fraction | |
KFold | Integer greater than 1 | |
Example: HyperparameterOptimizationOptions=struct(UseParallel=true)
Output Arguments
Trained kernel regression model, returned as a RegressionKernel model object or RegressionPartitionedKernel cross-validated model
object.
If you set any of the name-value pair arguments
CrossVal, CVPartition,
Holdout, KFold, or
Leaveout, then Mdl is a
RegressionPartitionedKernel cross-validated model.
Otherwise, Mdl is a RegressionKernel
model.
To reference properties of Mdl, use dot notation. For
example, enter Mdl.NumExpansionDimensions in the Command
Window to display the number of dimensions of the expanded space.
If you specify OptimizeHyperparameters and
set the ConstraintType and ConstraintBounds options of
HyperparameterOptimizationOptions, then Mdl is an
N-by-1 cell array of model objects, where N is equal
to the number of rows in ConstraintBounds. If none of the optimization
problems yields a feasible model, then each cell array value is [].
Note
Unlike other regression models, and for economical memory usage, a
RegressionKernel model object does not store the
training data or training process details (for example, convergence
history).
Aggregate optimization results for multiple optimization problems, returned as an AggregateBayesianOptimization object. To return
AggregateOptimizationResults, you must specify
OptimizeHyperparameters and
HyperparameterOptimizationOptions. You must also specify the
ConstraintType and ConstraintBounds
options of HyperparameterOptimizationOptions. For an example that
shows how to produce this output, see Hyperparameter Optimization with Multiple Constraint Bounds.
Optimization details, returned as a structure array including fields described in this table. The fields contain final values or name-value pair argument specifications.
| Field | Description |
|---|---|
Solver | Objective function minimization technique:
|
LossFunction | Loss function. Either mean squared error (MSE) or
epsilon-insensitive, depending on the type of linear
regression model. See Learner. |
Lambda | Regularization term strength. See
Lambda. |
BetaTolerance | Relative tolerance on the linear coefficients and the
bias term. See BetaTolerance. |
GradientTolerance | Absolute gradient tolerance. See
GradientTolerance. |
ObjectiveValue | Value of the objective function when optimization terminates. The regression loss plus the regularization term compose the objective function. |
GradientMagnitude | Infinite norm of the gradient vector of the objective
function when optimization terminates. See
GradientTolerance. |
RelativeChangeInBeta | Relative changes in the linear coefficients and the bias
term when optimization terminates. See
BetaTolerance. |
FitTime | Elapsed, wall-clock time (in seconds) required to fit the model to the data. |
History | History of optimization information. This field also
includes the optimization information from training
Mdl. This field is empty
([]) if you specify
'Verbose',0. For details, see
Verbose and Algorithms. |
To access fields, use dot notation. For example, to access the vector of
objective function values for each iteration, enter
FitInfo.ObjectiveValue in the Command Window.
If you specify
OptimizeHyperparameters and set the
ConstraintType and
ConstraintBounds options of
HyperparameterOptimizationOptions, then
Fitinfo is an N-by-1 cell array of
structure arrays, where N is equal to the number of rows
in ConstraintBounds.
Examine the information provided by FitInfo to assess
whether convergence is satisfactory.
Cross-validation optimization of hyperparameters, returned as a BayesianOptimization object, an AggregateBayesianOptimization object, or a table of hyperparameters and
associated values. The output is nonempty when
OptimizeHyperparameters has a value other than
"none".
If you set the ConstraintType and
ConstraintBounds options in
HyperparameterOptimizationOptions, then
HyperparameterOptimizationResults is an AggregateBayesianOptimization object. Otherwise, the value of
HyperparameterOptimizationResults depends on the value of the
Optimizer option in
HyperparameterOptimizationOptions.
Value of Optimizer Option | Value of HyperparameterOptimizationResults |
|---|---|
"bayesopt" (default) | Object of class BayesianOptimization |
"gridsearch" or "randomsearch" | Table of hyperparameters used, observed objective function values (cross-validation loss), and rank of observations from lowest (best) to highest (worst) |
More About
Random feature expansion, such as Random Kitchen Sinks [1] or Fastfood [2], is a scheme to approximate Gaussian kernels of the kernel regression algorithm for big data in a computationally efficient way. Random feature expansion is more practical for big data applications that have large training sets, but can also be applied to smaller data sets that fit in memory.
After mapping the predictor data into a high-dimensional space, the kernel regression algorithm searches for an optimal function that deviates from each response data point (yi) by values no greater than the epsilon margin (ε).
Some regression problems cannot be described adequately using a linear model. In such cases, obtain a nonlinear regression model by replacing the dot product x1x2′ with a nonlinear kernel function , where xi is the ith observation (row vector) and φ(xi) is a transformation that maps xi to a high-dimensional space (called the “kernel trick”). However, evaluating G(x1,x2), the Gram matrix, for each pair of observations is computationally expensive for a large data set (large n).
The random feature expansion scheme finds a random transformation so that its dot product approximates the Gaussian kernel. That is,
where T(x) maps x in to a high-dimensional space (). The Random Kitchen Sinks [1] scheme uses the random transformation
where is a sample drawn from and σ is a kernel scale. This scheme requires O(mp) computation and storage. The Fastfood [2] scheme introduces
another random basis V instead of Z using Hadamard
matrices combined with Gaussian scaling matrices. This random basis reduces computation cost
to O(mlogp) and reduces storage to O(m).
You can specify values for m and σ, using the
NumExpansionDimensions and KernelScale
name-value pair arguments of fitrkernel, respectively.
The fitrkernel function uses the Fastfood scheme for random feature
expansion and uses linear regression to train a Gaussian kernel regression model. Unlike
solvers in the fitrsvm function, which require computation of the
n-by-n Gram matrix, the solver in
fitrkernel only needs to form a matrix of size
n-by-m, with m typically much
less than n for big data.
A box constraint is a parameter that controls the maximum penalty imposed on observations that lie outside the epsilon margin (ε), and helps to prevent overfitting (regularization). Increasing the box constraint can lead to longer training times.
The box constraint (C) and the regularization term strength (λ) are related by C = 1/(λn), where n is the number of observations.
Tips
Standardizing predictors before training a model can be helpful.
You can standardize training data and scale test data to have the same scale as the training data by using the
normalizefunction.Alternatively, use the
Standardizename-value argument to standardize the numeric predictors before training. The returned model includes the predictor means and standard deviations in itsMuandSigmaproperties, respectively. (since R2023b)
After training a model, you can generate C/C++ code that predicts responses for new data. Generating C/C++ code requires MATLAB Coder™. For details, see Introduction to Code Generation.
Algorithms
fitrkernel minimizes the regularized objective function using a Limited-memory Broyden-Fletcher-Goldfarb-Shanno (LBFGS) solver with ridge (L2) regularization. To find the type of LBFGS solver used for training, type FitInfo.Solver in the Command Window.
'LBFGS-fast'— LBFGS solver.'LBFGS-blockwise'— LBFGS solver with a block-wise strategy. Iffitrkernelrequires more memory than the value ofBlockSizeto hold the transformed predictor data, then the function uses a block-wise strategy.'LBFGS-tall'— LBFGS solver with a block-wise strategy for tall arrays.
When fitrkernel uses a block-wise strategy, it implements LBFGS by
distributing the calculation of the loss and gradient among different parts of the data at
each iteration. Also, fitrkernel refines the initial estimates of the
linear coefficients and the bias term by fitting the model locally to parts of the data and
combining the coefficients by averaging. If you specify 'Verbose',1, then
fitrkernel displays diagnostic information for each data pass and
stores the information in the History field of
FitInfo.
When fitrkernel does not use a block-wise strategy, the initial estimates are zeros. If you specify 'Verbose',1, then fitrkernel displays diagnostic information for each iteration and stores the information in the History field of FitInfo.
References
[1] Rahimi, A., and B. Recht. “Random Features for Large-Scale Kernel Machines.” Advances in Neural Information Processing Systems. Vol. 20, 2008, pp. 1177–1184.
[2] Le, Q., T. Sarlós, and A. Smola. “Fastfood — Approximating Kernel Expansions in Loglinear Time.” Proceedings of the 30th International Conference on Machine Learning. Vol. 28, No. 3, 2013, pp. 244–252.
[3] Huang, P. S., H. Avron, T. N. Sainath, V. Sindhwani, and B. Ramabhadran. “Kernel methods match Deep Neural Networks on TIMIT.” 2014 IEEE International Conference on Acoustics, Speech and Signal Processing. 2014, pp. 205–209.
Extended Capabilities
The
fitrkernel function supports tall arrays with the following usage
notes and limitations:
fitrkerneldoes not support talltabledata.Some name-value pair arguments have different defaults compared to the default values for the in-memory
fitrkernelfunction. Supported name-value pair arguments, and any differences, are:'BoxConstraint''Epsilon''NumExpansionDimensions''KernelScale''Lambda''Learner''Verbose'— Default value is1.'BlockSize''RandomStream''ResponseTransform''Weights'— Value must be a tall array.'BetaTolerance'— Default value is relaxed to1e–3.'GradientTolerance'— Default value is relaxed to1e–5.'HessianHistorySize''IterationLimit'— Default value is relaxed to20.'OptimizeHyperparameters''HyperparameterOptimizationOptions'— For cross-validation, tall optimization supports only'Holdout'validation. By default, the software selects and reserves 20% of the data as holdout validation data, and trains the model using the rest of the data. You can specify a different value for the holdout fraction by using this argument. For example, specify'HyperparameterOptimizationOptions',struct('Holdout',0.3)to reserve 30% of the data as validation data.
If
'KernelScale'is'auto', thenfitrkerneluses the random stream controlled bytallrngfor subsampling. For reproducibility, you must set a random number seed for both the global stream and the random stream controlled bytallrng.If
'Lambda'is'auto', thenfitrkernelmight take an extra pass through the data to calculate the number of observations inX.fitrkerneluses a block-wise strategy. For details, see Algorithms.
For more information, see Tall Arrays.
To perform parallel hyperparameter optimization, use the UseParallel=true
option in the HyperparameterOptimizationOptions name-value argument in
the call to the fitrkernel function.
For more information on parallel hyperparameter optimization, see Parallel Bayesian Optimization.
For general information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
fitrkernelfits the model on a GPU if at least one of the following applies:The input argument
Xis agpuArrayobject.The input argument
Yis agpuArrayobject.The input argument
TblcontainsgpuArraypredictor or response variables.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2018afitrkernel fully supports GPU arrays.
fitrkernel defaults to serial hyperparameter optimization when
HyperparameterOptimizationOptions includes
UseParallel=true and the software cannot open a parallel pool.
In previous releases, the software issues an error under these circumstances.
Starting in R2023b, fitrkernel supports the standardization
of numeric predictors. That is, you can specify the Standardize
value as true to center and scale each numeric predictor variable
by the corresponding column mean and standard deviation. The software does not
standardize the categorical predictors.
You can also optimize the Standardize hyperparameter by using
the OptimizeHyperparameters name-value argument. Unlike in
previous releases, when you specify "auto" as the
OptimizeHyperparameters value,
fitrkernel includes Standardize as an
optimizable hyperparameter.
See Also
bayesopt | bestPoint | fitrlinear | fitrsvm | loss | predict | RegressionKernel | resume | RegressionPartitionedKernel
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)