checkCollision
Check if robot is in collision
Syntax
Description
[
checks if the specified rigid body tree robot model isSelfColliding,selfSeparationDist,selfWitnessPts] = checkCollision(robot,config)robot is in
self-collision at the specified configuration config. Add collision
objects to the rigid body tree robot model using the addCollision function. The checkCollision function also
returns the closest separation distance selfSeparationDist and the
witness points selfWitnessPts as points on each body.
The function ignores adjacent bodies when checking for self-collisions.
[
checks if the specified rigid body tree robot model is in collision with itself or a
specified set of collision objects in the world isColliding,separationDist,witnessPts] = checkCollision(robot,config,worldObjects)worldObjects.
[___] = checkCollision(___,
specifies additional options using one or more name-value pair arguments in addition to
any of argument combinations from previous syntaxes.Name,Value)
Examples
Load a robot model and modify the collision meshes. Clear existing collision meshes, add simple collision object primitives, and check whether certain configurations are in collision.
Load Robot Model
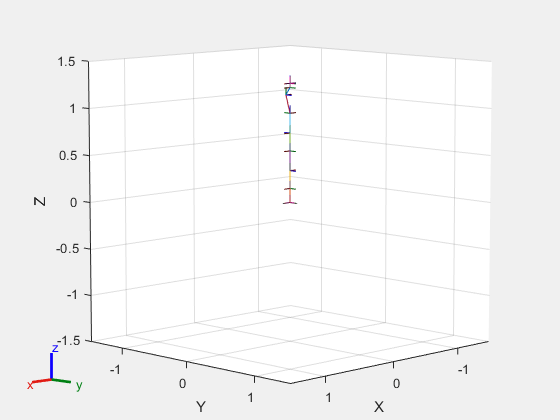
Load a preconfigured robot model into the workspace using the loadrobot function. This model already has collision meshes specified for each body. Iterate through all the rigid body elements and clear the existing collision meshes. Confirm that the existing meshes are gone.
robot = loadrobot("kukaIiwa7",DataFormat="column"); for i = 1:robot.NumBodies clearCollision(robot.Bodies{i}) end show(robot,Collisions="on",Visuals="off");
Add Collision Cylinders
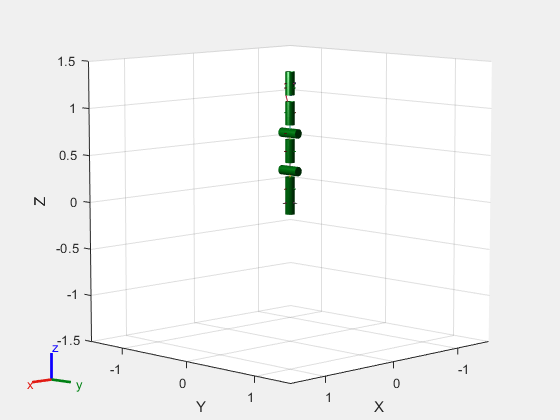
Iteratively add a blue collision cylinder to each body and set the transparency to opaque. Skip some bodies for this specific model, as they overlap and always collide with the end effector (body 10).
collisionObj = collisionCylinder(0.05,0.25); for i = 1:robot.NumBodies if i > 6 && i < 10 % Skip these bodies. else addCollision(robot.Bodies{i}, collisionObj, FaceColor=[0 0 1], FaceAlpha=1) end end show(robot,Collisions="on",Visuals="off");
Check for Collisions
Generate a series of random configurations. Check whether the robot is in collision at each configuration. Visualize each configuration that has a collision.
figure rng(0) % Set random seed for repeatability. for i = 1:20 config = randomConfiguration(robot); isColliding = checkCollision(robot,config,SkippedSelfCollisions="parent"); if isColliding show(robot,config,Collisions="on",Visuals="off"); title("Collision Detected") else % Skip non-collisions. end end
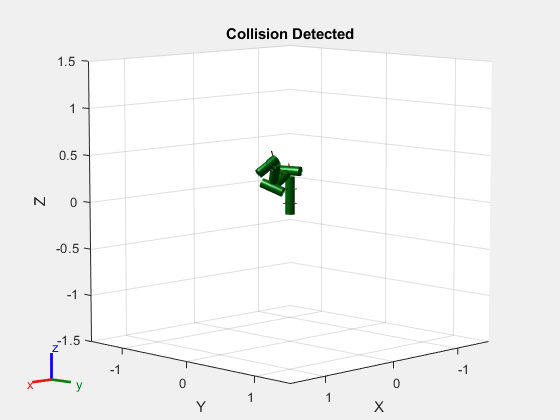
This example shows how to change which rigid body pairs are skipped during self-collision checking in rigid body trees using the SkippedSelfCollisions name-value argument for checkCollision.
Serial Manipulator Robot
Load a serial manipulator robot represented as a two joint rigid body tree. Since this robot does not have collision geometries, add some primitive collision geometries.
rbt2j = twoJointRigidBodyTree; P = [0.05 0.45]; % Geometry parameters for capsules T = trvec2tform([0.2 0 0]) * eul2tform([0 pi/2 0],"XYZ"); % Transformation parameters for capsules addCollision(rbt2j.Base,"cylinder",[0.075 0.1],trvec2tform([0 0 0.05])) addCollision(rbt2j.Bodies{1},"capsule",P,T) addCollision(rbt2j.Bodies{2},"capsule",P,T) addCollision(rbt2j.Bodies{3},"box",[0.2 0.05 0.2])
Visualize the robot with collisions on.
show(rbt2j,homeConfiguration(rbt2j),Collisions="on");
By default, SkippedSelfCollisions is "parent", so self-collision checking skips collisions between parent and child bodies. Check the parent and child bodies of body2.
body2 = rbt2j.Bodies{2};
rbt2j.BodyNamesans = 1×3 cell
{'body1'} {'body2'} {'tool'}
body2.Parent.Name
ans = 'body1'
body2.Children{1}.Nameans = 'tool'
This means that "body2" is not checked for collisions against "body1" or "tool".
List the body names of the robot. This shows that in the cell array, "body2", which is stored at index 2, is adjacent to both "body1" at index 1 and "tool" at index 3. Because the skipped collision pairs have not changed, the SkippedSelfCollisions name-value argument has no effect on the self-collision checking result for this robot.
rbt2j.BodyNames
ans = 1×3 cell
{'body1'} {'body2'} {'tool'}
Run collision checking with both SkippedSelfCollisions options to verify that the SkippedSelfCollisions name-value argument returns the same result for this robot.
checkCollision(rbt2j,homeConfiguration(rbt2j),SkippedSelfCollisions="parent")ans = logical
0
checkCollision(rbt2j,homeConfiguration(rbt2j),SkippedSelfCollisions="adjacent")ans = logical
0
Parallel Manipulator Robot
Use the exampleHelperCreate2ArmRBT example helper to create a parallel robot comprised of two one-joint arms.
rbt2arm = exampleHelperCreate2ArmRBT; show(rbt2arm,homeConfiguration(rbt2arm),Collisions="on"); axis padded

List the body names of the robot. The skipped body pairs formed by bodies of adjacent indices is similar to the serial manipulator but without body, "tool".
rbt2arm.BodyNames
ans = 1×2 cell
{'body1'} {'body2'}
Check the parent of body1 and the parent of body2. Each body forms a parent-child relationship with the base, even though they are at adjacent indices.
rbt2arm.Bodies{1}.Parent.Nameans = 'base'
rbt2arm.Bodies{2}.Parent.Nameans = 'base'
Run collision checking with both SkippedSelfCollisions options.
checkCollision(rbt2arm,homeConfiguration(rbt2arm),SkippedSelfCollisions="parent")ans = logical
0
checkCollision(rbt2arm,homeConfiguration(rbt2arm),SkippedSelfCollisions="adjacent")ans = logical
1
As expected, when skipping parent-child body pairs during self collision checks, checkCollision finds no self collisions, but does find a self collision between the "base" and "body2" when skipping body pairs of adjacent indices.
When you want to resolve all self-collisions in a robot model without checking them between bodies with a parent-child relationship or at adjacent indices, you can use the SkippedSelfCollisions name-value argument with the "parent" and "adjacent" values, respectively. However, if your robot model has one or more self-collisions that you can not resolve when using either "parent" or "adjacent", you can specify additional body pairs between which to skip collision checking.
Load an ABB IRB 1600 robot model into the workspace, with a data format of "row". This is an example of a robot model that contains an additional self-collision when you specify the SkippedSelfCollision name-value argument as "adjacent".
rbt = loadrobot("abbIrb1600",DataFormat="row"); config = homeConfiguration(rbt)
config = 1×6
0 0 0 0 0 0
Perform an exhaustive self-collision check, and skip self-collisions between bodies at adjacent indices
[isSelfColl1,sepDist1] = checkCollision(rbt,config,SkippedSelfCollisions="adjacent",Exhaustive="on");
You can determine which pairs of bodies are in collision by checking the separation distance matrix sepDist1 for NaN values. To make the separation distance matrix easier to read, convert the separation distance matrix to a table. Use the body names and base link name to label the rows and columns. The NaN values indicate that the self-collision is between link_4 and link_6..
bodynames = [rbt.BodyNames rbt.Base.Name]; collTable = array2table(sepDist1,VariableNames=bodynames,RowNames=bodynames)
collTable=8×8 table
link_1 link_2 link_3 link_4 link_5 link_6 tool0 base_link
_______ _______ _______ _______ _______ _______ _____ _________
link_1 Inf Inf 0.176 0.36201 0.55837 0.59557 Inf Inf
link_2 Inf Inf Inf 0.25321 0.49887 0.54104 Inf 0.25599
link_3 0.176 Inf Inf Inf 0.244 0.286 Inf 0.65488
link_4 0.36201 0.25321 Inf Inf Inf NaN Inf 0.7801
link_5 0.55837 0.49887 0.244 Inf Inf Inf Inf 0.91534
link_6 0.59557 0.54104 0.286 NaN Inf Inf Inf 0.95033
tool0 Inf Inf Inf Inf Inf Inf Inf Inf
base_link Inf 0.25599 0.65488 0.7801 0.91534 0.95033 Inf Inf
Check the value of isSelfColl1. The output argument returns 1 (true), indicating that the robot model has a self-collision between a pair of bodies that are not adjacent.
isSelfColl1
isSelfColl1 = logical
1
To skip checking for self-collision between link_4 and link_6, you must manually specify all of the body pairs for which to skip checking for self-collision. First, create a list of all the body names in the model, in order from the base link to the tool. Then, by combining the ordered list of rigid body names with an offset version of itself, create a two-column cell array that specifies each adjacent rigid body pair. Then add link_4 and link_6 as an additional skipped body pair.
adjbodynames = [rbt.Base.Name rbt.BodyNames]
adjbodynames = 1×8 cell
{'base_link'} {'link_1'} {'link_2'} {'link_3'} {'link_4'} {'link_5'} {'link_6'} {'tool0'}
skiplist = cell(rbt.NumBodies,2); for i = 1:rbt.NumBodies skiplist(i,:) = {adjbodynames{i}, adjbodynames{i+1}}; end skiplist = [skiplist; {'link_4','link_6'}]
skiplist = 8×2 cell
{'base_link'} {'link_1'}
{'link_1' } {'link_2'}
{'link_2' } {'link_3'}
{'link_3' } {'link_4'}
{'link_4' } {'link_5'}
{'link_5' } {'link_6'}
{'link_6' } {'tool0' }
{'link_4' } {'link_6'}
Perform self-collision checking, and note that checkCollision no longer indicates self-collision between link_4 and link_6.
[isSelfColl2,sepDist2] = checkCollision(rbt,config,SkippedSelfCollisions=skiplist,Exhaustive="on");
isSelfColl2isSelfColl2 = logical
0
array2table(sepDist2,VariableNames=bodynames,RowNames=bodynames)
ans=8×8 table
link_1 link_2 link_3 link_4 link_5 link_6 tool0 base_link
_______ _______ _______ _______ _______ _______ _____ _________
link_1 Inf Inf 0.176 0.36201 0.55837 0.59557 Inf Inf
link_2 Inf Inf Inf 0.25321 0.49887 0.54104 Inf 0.25599
link_3 0.176 Inf Inf Inf 0.244 0.286 Inf 0.65488
link_4 0.36201 0.25321 Inf Inf Inf Inf Inf 0.7801
link_5 0.55837 0.49887 0.244 Inf Inf Inf Inf 0.91534
link_6 0.59557 0.54104 0.286 Inf Inf Inf Inf 0.95033
tool0 Inf Inf Inf Inf Inf Inf Inf Inf
base_link Inf 0.25599 0.65488 0.7801 0.91534 0.95033 Inf Inf
Input Arguments
Rigid body tree robot model, specified as a rigidBodyTree object. To use the checkCollision function,
the DataFormat property of the
rigidBodyTree object must be either 'row' or
'column'.
Joint configuration of the rigid body tree, specified as an n-element numeric vector, where n is the number of nonfixed joints in the robot model. Each element of the vector is a specific joint position for a joint in the robot model.
Data Types: single | double
List of collision objects in the world, specified as a cell array of collision
objects with any combination of collisionBox, collisionCylinder, collisionSphere, and collisionMesh objects. The function assumes that the
Pose property of each object is relative to the base of the rigid
body tree robot model.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: Exhaustive="on" enables exhaustive checking for collisions
and causes the function to calculate all separation distances and witness
points.
Exhaustively check for all collisions, specified as the comma-separated pair
consisting of "Exhaustive" and "on" or
"off". By default, the function finds the first collision and
stops, returning the separation distances and witness points for incomplete checks as
Inf.
If this name-value pair argument is specified as "on", the
function instead continues checking for collisions until it has exhausted all
possibilities.
Data Types: char | string
Skip checking for robot self-collisions, specified as the comma-separated pair
consisting of "IgnoreSelfCollision" and "on" or
"off". When this argument is enabled, the function ignores
collisions between the collision objects of the rigid body tree robot model bodies and
other collision objects of the same model or its base.
This name-value pair argument affects the size of the
separationDist and witnessPts output
arguments.
Data Types: char | string
Body pairs skipped for checking self-collisions, specified as "parent",
"adjacent", or a p-by-2 cell array of
character vectors:
"parent"— Skip collision checking between child and parent bodies. See Skip Self-Collision Checking Between Parent and Adjacent Bodies for more information."adjacent"— Skip collision checking between bodies on adjacent indices. See Skip Self-Collision Checking Between Parent and Adjacent Bodies for more information.p-by-2 cell array of character vectors — Skip collision checking between specific body pairs. Each row specifies a pair of body names between which to skip self-collision checking. p is the number of body pairs to skip for self-collision checking. For more information, see Skip Self-Collision Checking Between Specific Body Pairs.
Data Types: char | string
Output Arguments
Self Collisions
Robot configuration is in self-collision returned as a logical 0
(false) or 1 (true). If the
function returns a value of true for this argument, that means that
one of the rigid body collision objects is touching another collision object in the
robot model. Add collision objects to your rigid body tree robot model using the
addCollision function.
Data Types: logical
Minimum separation distance between the bodies of the robot, returned as an (m+1) -by-(m+1) matrix, where m is the number of bodies. The final row and column correspond to the robot base. Units are in meters.
If a pair is in collision, the function returns the separation distance for the
associated element as NaN.
Data Types: double
Witness points between the robot bodies including the base, returned as an 3(m+1)-by-2(m+1) matrix, where m is the number of bodies. Witness points are the points on any two bodies that are closest to one another for a given configuration. The matrix takes the form:
The matrix is divided into 3-by-2 sections that represent the xyz-coordinates of witness point pairs in the form:
Each section corresponds to a separation distance in the
selfSeparationDist output matrix. Use these equations to
determine where the section of the selfWitnessPts matrix that
corresponds to a specific separation distance begins:
Where (Sr,Sc) is the index of a separation distance in the separation distance matrix and (Wr,Wc) is the index in the witness point matrix at which the corresponding witness points begin.
If a pair is in collision, the function returns each coordinate of the witness
points for that element as NaN.
Data Types: double
World Collisions
Robot configuration is in collision, returned as a two-element logical vector. The first element indicates whether the robot is in self-collision. The second element indicates whether the robot model is in collision with any world objects.
Data Types: logical
Minimum separation distance between the collision objected, returned as an (m+1)-by-(m+w+1) matrix, where m is the number of bodies and w is the number of world objects. The first m rows correspond to the robot bodies, where the (m+1)th row or column index corresponds to the base. The remaining w columns correspond to the world objects.
The matrix is divided into 3-by-2 sections that represent the xyz-coordinates of witness point pairs in the form:
Each section corresponds to a separation distance in the
separationDist output matrix. Use these equations to determine
where the section of the witnessPts matrix that corresponds to a
specific separation distance begins:
Where (Sr,Sc) is the index of a separation distance in the separation distance matrix and (Wr,Wc) is the index in the witness point matrix at which the corresponding witness points begin.
If a pair is in collision, the function returns each coordinate of the witness
points for that element as NaN.
If a pair is in collision, the function returns the separation distance as
NaN.
Dependencies
If you specify the "IgnoreSelfCollision" name-value pair
argument as "on", then the matrix does not contain values for the
distances between any given body and other bodies in the robot model.
Data Types: double
Witness points between collision objects, specified as a
3(m+1)-by-2(m+w+1) matrix,
where m is the number of bodies and w is the
number of world objects. Witness points are the points on any two bodies that are
closest to one another for a given configuration. The matrix takes the form:
[Wr1_1 Wr1_2 ... Wr1_(N+1) Wo1_1 Wo1_2 ... W1_M; Wr2_1 Wr2_2 ... Wr2_(N+1) Wo2_1 Wo2_2 ... W2_M; . . . . . . . . . . . . . . . . . . . . . . . . Wr(N+1)_1 Wr(N+1)_2 ... Wr(N+1)_(N+1) Wo(N+1)_1 Wo(N+1)_2 ... W(N+1)_M]
Each element in the above matrix is a 3-by-2 matrix that gives the nearest
[x y z]' points on the two corresponding bodies or world objects.
The final row and column correspond to the robot base.
Wr elements represent the witness points between the robot
bodies, selfWitnessPts. Woi_j elements represent
the witness points between the robot bodies and base, and the world objects. The
i indices correspond to the robot bodies, and the
j indices correspond to collision objects from the world object
list worldObjects.
If a pair is in collision, witness points for that element are returned as
NaN(3,2).
Dependencies
If the "IgnoreSelfCollision" name-value pair is set to
"on", then the matrix contains no Wr
elements.
Data Types: double
Extended Capabilities
Usage notes and limitations:
When creating the rigidBodyTree object, use the syntax that specifies the
MaxNumBodies as an upper bound for adding bodies to the robot model.
You must also specify the DataFormat property as a name-value pair. For
example:
robot = rigidBodyTree("MaxNumBodies",15,"DataFormat","row")
To minimize data usage, limit the upper bound to a number close to the expected number of bodies in the model. All data formats are supported for code generation. To use the dynamics functions, the data format must be set to "row" or "column".
The show and showdetails functions do not support code generation.
Version History
Introduced in R2020bYou can now select which body pairs to skip in self-collision checks by specifying the
SkippedSelfCollisions name-value argument as a cell array of body
pairs. For more information, see the SkippedSelfCollisions name-value argument.
You can now specify self-collision checking behavior for a rigid body tree robot model
by using the SkippedSelfCollisions name-value argument. Specify
SkippedSelfCollisions as "parent" or
"adjacent":
"parent"— Collision checking ignores self-collisions between parent and child rigid bodies."adjacent"— Collision checking ignores self -collisions between rigid bodies of adjacent indices.
As of R2022b, the default behavior of collision checking is to ignore self-collisions
between parent and child rigid bodies. In previous releases, the default behavior of
self-collision checking was to ignore self-collisions between adjacent rigid bodies. To
instead ignore self-collisions between rigid bodies of adjacent indices, specify
SkippedSelfCollisions as "adjacent".
See Skip Self-Collision Checking Between Parent and Adjacent Bodies for more information about how to use this name-value argument.
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)