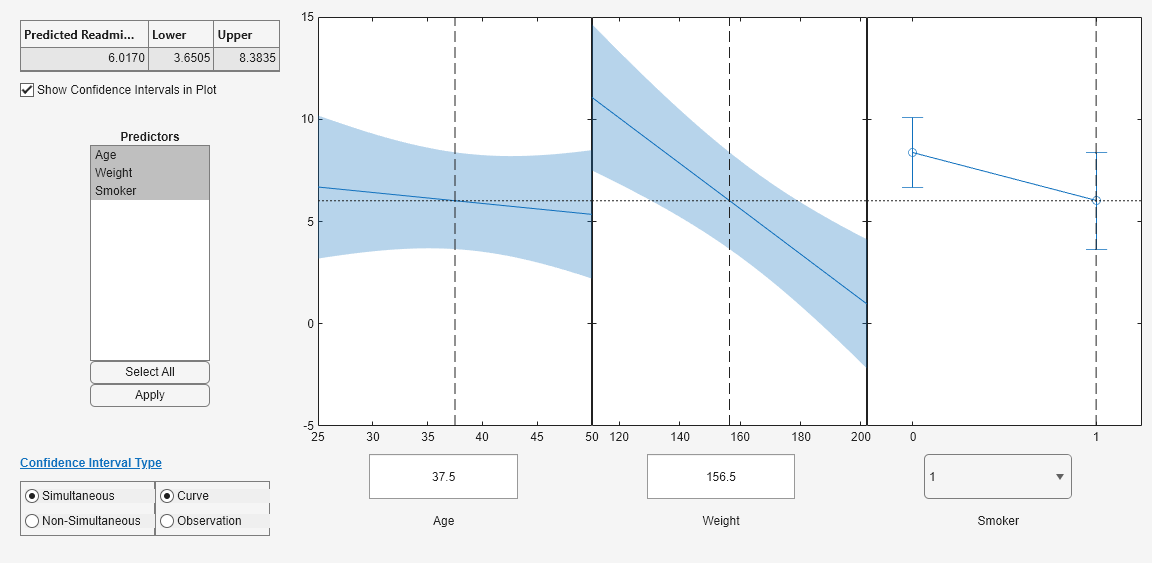
plotSlice
Description
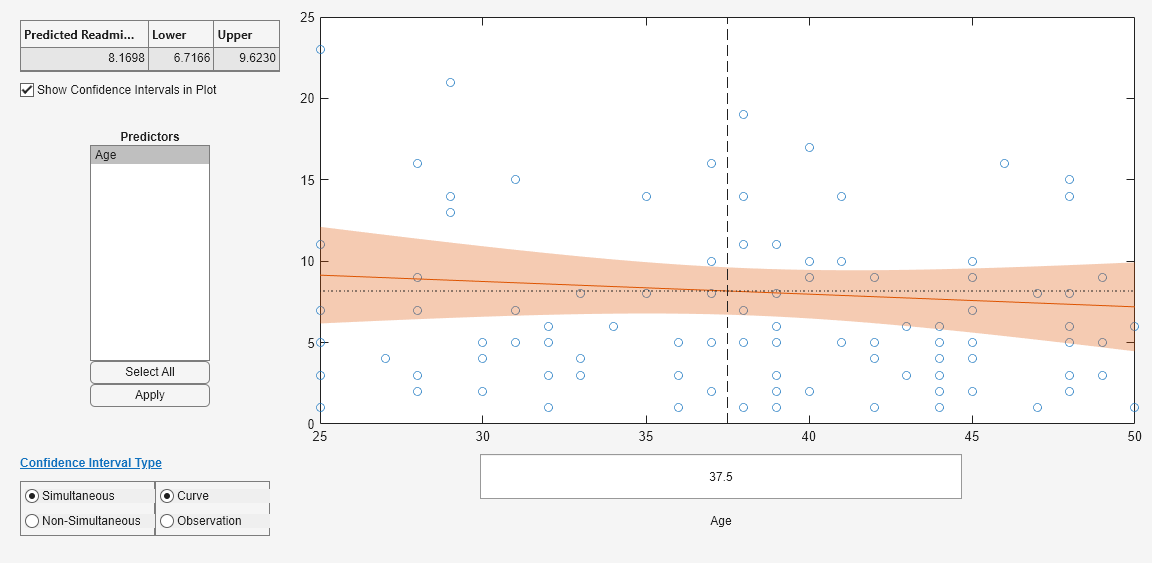
plotSlice( creates a figure containing one
or more plots, each representing a slice through the regression surface predicted by
mdl)mdl. Each plot shows the fitted response values as a function of a
single predictor variable, with the other predictor variables held constant.
plotSlice also displays the 95% confidence bounds for the response
values. You can specify the type of confidence bounds, and select which predictors to use
for each slice plot. For more information, see Tips.
Examples
Input Arguments
Tips
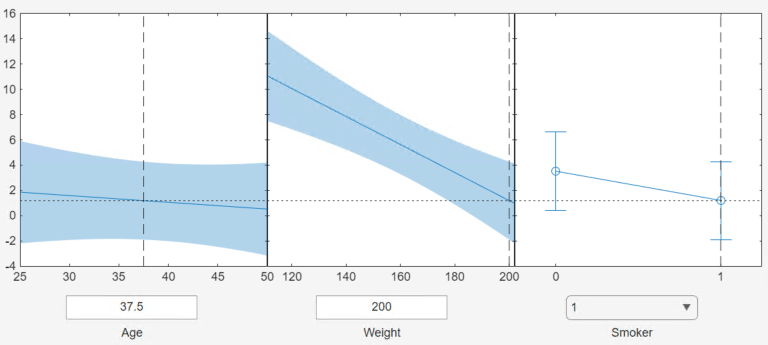
Use the Confidence Interval Type options in the figure window to specify the type of confidence bounds. You can specify simultaneous or nonsimultaneous, and curve or observation.
Simultaneous or Non-Simultaneous
Simultaneous (default) —
plotSlicecomputes confidence bounds for the curve of the response values using Scheffé's method. The range between the upper and lower confidence bounds contains the curve consisting of true response values with 95% confidence.Non-Simultaneous —
plotSlicecomputes confidence bounds for the response value at each observation. The confidence interval for a response value at a specific predictor value contains the true response value with 95% confidence.
With simultaneous bounds, the entire curve of true response values is within the bounds at high confidence. By contrast, non-simultaneous bounds require only the response value at a single predictor value to be within the bounds at high confidence. Therefore, simultaneous bounds are wider than non-simultaneous bounds.
If you do not want the plot to display confidence bounds, you can clear the Show Confidence Intervals in Plot box.
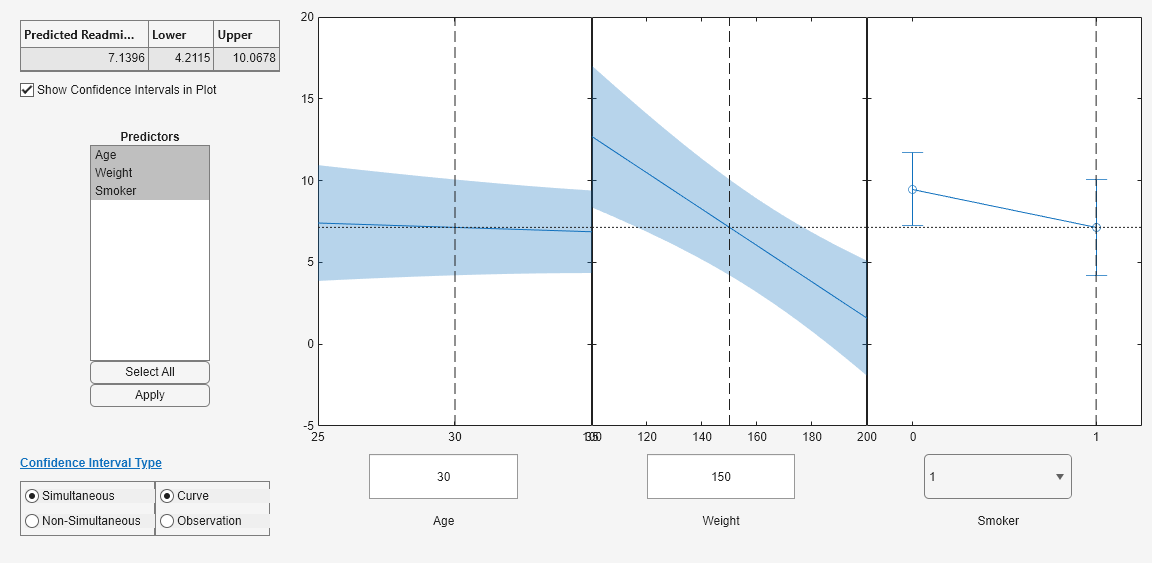
Curve or Observation
A regression model for the predictor variables X and the response variable y has the form
y = f(X) + ε,
where f is a function of X and ε is a random noise term.
Curve (default) —
plotSlicepredicts confidence bounds for the fitted responses f(X).Observation —
plotSlicepredicts confidence bounds for the response observations y.
The bounds for y are wider than the bounds for f(X) because of the additional variability of the noise term.
Use the Predictors options to select which predictors to use for each slice plot. If the regression model
mdlincludes more than eight predictors,plotSlicecreates plots for the first five predictors by default.
Alternative Functionality
Use
predictto return the predicted response values and confidence bounds. You can also specify the confidence level for confidence bounds by using theAlphaname-value argument of thepredictfunction. Note thatpredictfinds nonsimultaneous bounds by default, whereasplotSlicefinds simultaneous bounds by default.A
CensoredLinearModelobject provides multiple plotting functions.When verifying a model, use
plotResidualsto analyze the residuals of the model.After fitting a model, use
plotPartialDependenceto understand the effect of a particular predictor.
Version History
Introduced in R2025a