How Solvers Compute in Parallel
Parallel Processing Types in Global Optimization Toolbox
Parallel processing is an attractive way to speed optimization algorithms. To use
parallel processing, you must have a Parallel Computing Toolbox™ license, and have a parallel worker pool
(parpool). For more information, see How to Use Parallel Processing in Global Optimization Toolbox.
Global Optimization Toolbox solvers use parallel computing in various ways.
| Solver | Parallel? | Parallel Characteristics |
|---|---|---|
| × | No parallel functionality. However, fmincon
can use parallel gradient estimation when run in GlobalSearch. See Using Parallel Computing in Optimization Toolbox. |
|
| Start points distributed to multiple processors. From these points, local solvers run to completion. For more details, see MultiStart and How to Use Parallel Processing in Global Optimization Toolbox. |
For fmincon, no parallel gradient estimation
with parallel MultiStart. | ||
|
| Population evaluated in parallel, which occurs once per iteration. For more details, see Genetic Algorithm and How to Use Parallel Processing in Global Optimization Toolbox. |
| No vectorization of fitness or constraint functions. | ||
|
| Population evaluated in parallel, which occurs once per iteration. For more details, see Particle Swarm and How to Use Parallel Processing in Global Optimization Toolbox. |
| No vectorization of objective or constraint functions. | ||
|
| Poll points evaluated in parallel, which occurs once per iteration. For more details, see Pattern Search and How to Use Parallel Processing in Global Optimization Toolbox. |
| No vectorization of objective or constraint functions. | ||
| × | No parallel functionality. However,
simulannealbnd can use a hybrid function
that runs in parallel. See Simulated Annealing. |
|
| Search points evaluated in parallel. |
| No vectorization of objective or constraint functions. |
In addition, several solvers have hybrid functions that run after they finish.
Some hybrid functions can run in parallel. Also, most
patternsearch search methods can run in parallel. For more
information, see Parallel Search Functions or Hybrid Functions.
How Toolbox Functions Distribute Processes
parfor Characteristics and Caveats
No Nested parfor Loops. Most solvers employ the Parallel Computing Toolbox
parfor (Parallel Computing Toolbox) function to perform
parallel computations. Two solvers, surrogateopt and
paretosearch, use parfeval (Parallel Computing Toolbox) instead.
Note
parfor does not work in parallel when called from
within another parfor loop.
Note
The documentation recommends not to use parfor or
parfeval when calling Simulink®; see Using sim Function Within parfor (Simulink). Therefore, you might
encounter issues when optimizing a Simulink simulation in parallel using a solver's built-in parallel functionality. For
an example showing how to optimize a Simulink model with several Global Optimization Toolbox solvers, see Optimize Simulink Model in Parallel.
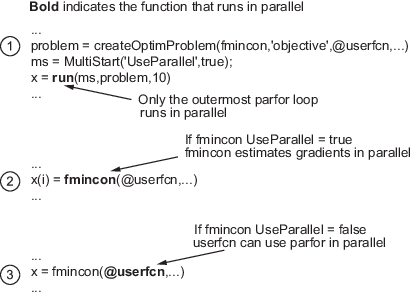
Suppose, for example, your objective function userfcn
calls parfor, and you want to call
fmincon using MultiStart and parallel processing. Suppose also that the
conditions for parallel gradient evaluation of fmincon
are satisfied, as given in Parallel Optimization Functionality. The figure When parfor Runs In Parallel shows
three cases:
The outermost loop is parallel
MultiStart. Only that loop runs in parallel.The outermost
parforloop is infmincon. Onlyfminconruns in parallel.The outermost
parforloop is inuserfcn. In this case,userfcncan useparforin parallel.
When parfor Runs In Parallel
Parallel Random Numbers Are Not Reproducible. Random number sequences in MATLAB® are pseudorandom, determined from a seed, or an initial setting. Parallel computations use seeds that are not necessarily controllable or reproducible. For example, each instance of MATLAB has a default global setting that determines the current seed for random sequences.
For patternsearch, if you select MADS as a poll or
search method, parallel pattern search does not have reproducible runs. If
you select the genetic algorithm or Latin hypercube as search methods,
parallel pattern search does not have reproducible runs.
For ga and gamultiobj, parallel
population generation gives nonreproducible results.
MultiStart is different. You
can have reproducible runs from parallel MultiStart. Runs are reproducible because
MultiStart generates pseudorandom
start points locally, and then distributes the start points to parallel
processors. Therefore, the parallel processors do not use random numbers.
For more details, see Parallel Processing and Random Number Streams.
Limitations and Performance Considerations. More caveats related to parfor appear in Parallel for-Loops (parfor) (Parallel Computing Toolbox).
For information on factors that affect the speed of parallel computations, and factors that affect the results of parallel computations, see Improving Performance with Parallel Computing. The same considerations apply to parallel computing with Global Optimization Toolbox functions.
MultiStart
MultiStart can automatically distribute a
problem and start points to multiple processes or processors. The problems run
independently, and MultiStart combines the
distinct local minima into a vector of GlobalOptimSolution objects. MultiStart uses parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the
UseParallelproperty totruein theMultiStartobject:ms = MultiStart('UseParallel',true);
When these conditions hold, MultiStart
distributes a problem and start points to processes or processors one at a time.
The algorithm halts when it reaches a stopping condition or runs out of start
points to distribute. If the MultiStart
Display property is 'iter', then MultiStart displays:
Running the local solvers in parallel.
For an example of parallel MultiStart, see
Parallel MultiStart.
Implementation Issues in Parallel MultiStart. fmincon cannot estimate gradients in parallel when
used with parallel MultiStart. This lack
of parallel gradient estimation is due to the limitation of
parfor described in No Nested parfor Loops.
fmincon can take longer to estimate gradients in
parallel rather than in serial. In this case, using MultiStart with parallel gradient estimation in
fmincon amplifies the slowdown. For example,
suppose the ms
MultiStart object has
UseParallel set to false. Suppose
fmincon takes 1 s longer to solve
problem with
problem.options.UseParallel set to
true. Then run(ms,problem,200)
takes 200 s longer than the same run with
problem.options.UseParallel set to
false
Note
When executing serially, parfor loops run slower
than for loops. Therefore, for best performance,
set your local solver UseParallel option to
false when the MultiStart
UseParallel property is
true.
Note
Even when running in parallel, a solver occasionally calls the objective and nonlinear constraint functions serially on the host machine. Therefore, ensure that your functions have no assumptions about whether they are evaluated in serial and parallel.
GlobalSearch
GlobalSearch does not distribute a problem
and start points to multiple processes or processors. However, when GlobalSearch runs the fmincon
local solver, fmincon can estimate gradients by parallel
finite differences. fmincon uses parallel computing when
you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the
UseParalleloption totruewithoptimoptions. Set this option in theproblemstructure:opts = optimoptions(@fmincon,'UseParallel',true,'Algorithm','sqp'); problem = createOptimProblem('fmincon','objective',@myobj,... 'x0',startpt,'options',opts);
For more details, see Using Parallel Computing in Optimization Toolbox.
Pattern Search
patternsearch can automatically distribute the evaluation
of objective and constraint functions associated with the points in a pattern to
multiple processes or processors. patternsearch uses
parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the following options using
optimoptions:UseCompletePollistrue.UseVectorizedisfalse(default).UseParallelistrue.
When these conditions hold, the solver computes the objective function and
constraint values of the pattern search in parallel during a poll. Furthermore,
patternsearch overrides the setting of the
Cache option, and uses the default
'off' setting.
Beginning in R2019a, when you set the
UseParallel option to true,
patternsearch internally overrides the
UseCompletePoll setting to true so that the function
polls in parallel.
Note
Even when running in parallel, patternsearch
occasionally calls the objective and nonlinear constraint functions serially
on the host machine. Therefore, ensure that your functions have no
assumptions about whether they are evaluated in serial or parallel.
Parallel Search Function. patternsearch can optionally call a search function
at each iteration. The search is parallel when you:
Set
UseCompleteSearchtotrue.Do not set the search method to
@searchneldermeadorcustom.Set the search method to a
patternsearchpoll method or Latin hypercube search, and setUseParalleltotrue.Or, if you set the search method to
ga, create a search method option withUseParallelset totrue.
Implementation Issues in Parallel Pattern Search. The limitations on patternsearch options, listed in
Pattern Search, arise partly from the
limitations of parfor, and partly from the nature of
parallel processing:
Cacheis overridden to be'off'—patternsearchimplementsCacheas a persistent variable.parfordoes not handle persistent variables, because the variable could have different settings at different processors.UseCompletePollistrue—UseCompletePolldetermines whether a poll stops as soon aspatternsearchfinds a better point. When searching in parallel,parforschedules all evaluations simultaneously, andpatternsearchcontinues after all evaluations complete.patternsearchcannot halt evaluations after they start.Beginning in R2019a, when you set the
UseParalleloption totrue,patternsearchinternally overrides theUseCompletePollsetting totrueso that the function polls in parallel.UseVectorizedisfalse—UseVectorizeddetermines whetherpatternsearchevaluates all points in a pattern with one function call in a vectorized fashion. IfUseVectorizedistrue,patternsearchdoes not distribute the evaluation of the function, so does not useparfor.
Genetic Algorithm
ga and gamultiobj can automatically
distribute the evaluation of objective and nonlinear constraint functions
associated with a population to multiple processors. ga
uses parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the following options using
optimoptions:UseVectorizedisfalse(default).UseParallelistrue.
When these conditions hold, ga computes the objective
function and nonlinear constraint values of the individuals in a population in
parallel.
Note
Even when running in parallel, ga occasionally calls
the fitness and nonlinear constraint functions serially on the host machine.
Therefore, ensure that your functions have no assumptions about whether they
are evaluated in serial or parallel.
Implementation Issues in Parallel Genetic Algorithm. The limitations on options, listed in Genetic Algorithm, arise partly from limitations of
parfor, and partly from the nature of parallel processing:
UseVectorizedisfalse—UseVectorizeddetermines whethergaevaluates an entire population with one function call in a vectorized fashion. IfUseVectorizedistrue,gadoes not distribute the evaluation of the function, so does not useparfor.
ga can have a hybrid function that runs after it
finishes; see Hybrid Scheme in the Genetic Algorithm. If you want the
hybrid function to take advantage of parallel computation, set its options
separately so that UseParallel is
true. If the hybrid function is
patternsearch, set
UseCompletePoll to true so that
patternsearch runs in parallel.
If the hybrid function is fmincon, set the following
options with optimoptions to have parallel gradient estimation:
GradObjmust not be'on'— it can be'off'or[].Or, if there is a nonlinear constraint function,
GradConstrmust not be'on'— it can be'off'or[].
To find out how to write options for the hybrid function, see Parallel Hybrid Functions.
Parallel Computing with gamultiobj
Parallel computing with gamultiobj works almost the same
as with ga. For detailed information, see Genetic Algorithm.
The difference between parallel computing with gamultiobj
and ga has to do with the hybrid function.
gamultiobj allows only one hybrid function,
fgoalattain. This function optionally runs after
gamultiobj finishes its run. Each individual in the
calculated Pareto frontier, that is, the final population found by
gamultiobj, becomes the starting point for an
optimization using fgoalattain. These optimizations run in
parallel. The number of processors performing these optimizations is the smaller
of the number of individuals and the size of your
parpool.
For fgoalattain to run in parallel, set its options
correctly:
fgoalopts = optimoptions(@fgoalattain,'UseParallel',true) gaoptions = optimoptions('ga','HybridFcn',{@fgoalattain,fgoalopts});
gamultiobj with gaoptions, and
fgoalattain runs in parallel. For more information
about setting the hybrid function, see Hybrid Function Options.gamultiobj calls fgoalattain using a
parfor loop, so fgoalattain does
not estimate gradients in parallel when used as a hybrid function with
gamultiobj. For more information, see No Nested parfor Loops.
Particle Swarm
particleswarm can automatically distribute the evaluation
of the objective function associated with a population to multiple processors.
particleswarm uses parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the following options using
optimoptions:UseVectorizedisfalse(default).UseParallelistrue.
When these conditions hold, particleswarm computes the
objective function of the particles in a population in parallel.
Note
Even when running in parallel, particleswarm
occasionally calls the objective function serially on the host machine.
Therefore, ensure that your objective function has no assumptions about
whether it is evaluated in serial or parallel.
Implementation Issues in Parallel Particle Swarm Optimization. The limitations on options, listed in Particle Swarm, arise partly from limitations of
parfor, and partly from the nature of parallel processing:
UseVectorizedisfalse—UseVectorizeddetermines whetherparticleswarmevaluates an entire population with one function call in a vectorized fashion. IfUseVectorizedistrue,particleswarmdoes not distribute the evaluation of the function, so does not useparfor.
particleswarm can have a hybrid function that runs
after it finishes; see Hybrid Scheme in the Genetic Algorithm. If you want the hybrid function to
take advantage of parallel computation, set its options separately so that
UseParallel is true. If the hybrid
function is patternsearch, set
UseCompletePoll to true so that
patternsearch runs in parallel.
If the hybrid function is fmincon, set the
GradObj option to 'off' or
[] with optimoptions to have
parallel gradient estimation.
To find out how to write options for the hybrid function, see Parallel Hybrid Functions.
Simulated Annealing
simulannealbnd does not run in parallel automatically.
However, it can call hybrid functions that take advantage of parallel computing.
To find out how to write options for the hybrid function, see Parallel Hybrid Functions.
Pareto Search
paretosearch can automatically distribute the evaluation
of the objective function associated with a population to multiple processors.
paretosearch uses parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the following option using
optimoptions:UseParallelistrue.
When these conditions hold, paretosearch computes the
objective function of the particles in a population in parallel.
Note
Even when running in parallel, paretosearch
occasionally calls the objective function serially on the host machine.
Therefore, ensure that your objective function has no assumptions about
whether it is evaluated in serial or parallel.
For algorithmic details, see Modifications for Parallel Computation and Vectorized Function Evaluation.
Surrogate Optimization
surrogateopt can automatically distribute the evaluation
of the objective function associated with a population to multiple processors.
surrogateopt uses parallel computing when you:
Have a license for Parallel Computing Toolbox software.
Enable parallel computing with
parpool, a Parallel Computing Toolbox function.Set the following option using
optimoptions:UseParallelistrue.
When these conditions hold, surrogateopt computes the
objective function of the particles in a population in parallel.
Note
Even when running in parallel, surrogateopt
occasionally calls the objective function serially on the host machine.
Therefore, ensure that your objective function has no assumptions about
whether it is evaluated in serial or parallel.
For algorithmic details, see Parallel surrogateopt Algorithm.