bioinfo.pipeline.block.UserFunction
Description
A UserFunction block enables you to use any existing or custom
function as a block in your pipeline, similar to any other built-in blocks.
Creation
Syntax
Description
ufBlock = bioinfo.pipeline.block.UserFunctionUserFunction block.
ufBlock = bioinfo.pipeline.block.UserFunction(fcn)UserFunction block from a custom function
fcn, which can be a function handle, name of an existing or custom
function, or function signature string.
ufBlock = bioinfo.pipeline.block.UserFunction(fcn,Name=Value)fcn must be a function handle or name of a function.
Input Arguments
Custom function, specified as a function handle, string or character vector representing the name of a function.
Data Types: char | string | function_handle
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: blosumBlock = UserFunction(@blosum62,
OutputArguments="Matrix") specifies to create a UserFunction
block for the blosum62 function using the string
"Matrix" as the name of the block output port.
Names of the required positional input arguments for the custom function
fcn, specified as a string, character vector, string vector,
or cell array of character vectors. The order of arguments specified in this
property is the same order used to call the underlying function
fcn. The specified names are used as the names of required
input ports of the block.
The corresponding input ports of the UserFunction block have
the Required property set to true to indicate that these ports
are required and must be satisfied.
Names of the output arguments returned by the custom function
fcn, specified as a string, character vector, string vector,
or cell array of character vectors.
Names of the optional name-value arguments for the custom function
fcn, specified as a string, character vector, string vector,
or cell array of character vectors.
The corresponding input ports of the UserFunction block have
the Required property set to false to indicate that these ports
are optional.
Properties
Function to handle errors from the run
method of the block, specified as a function handle. The handle specifies the function to call
if the run method encounters an error within a pipeline. For the pipeline to continue after a
block fails, ErrorHandler must return a structure that is compatible with
the output ports of the block. The error handling function is called with the following two inputs:
Structure with these fields:
Field Description identifier Identifier of the error that occurred message Text of the error message index Linear index indicating which block process failed in the parallel run. By default, the index is 1 because there is only one run per block. For details on how block inputs can be split across different dimensions for multiple run calls, see Bioinformatics Pipeline SplitDimension. Input structure passed to the
runmethod when it fails
Data Types: function_handle
Function to evaluate when you run the block, specified as a function handle, string scalar, or character vector that represents the name of any custom function.
When you call the run method with an input structure, it
converts the input structure to positional and name-value arguments as determined by the
Signature property and runs the specified function with those
converted inputs. For examples, see Create UserFunction Blocks For MATLAB Functions.
Data Types: char | string | function_handle
This property is read-only.
Input ports of the block, specified as a structure. The field
names of the structure are the names of the block input ports, and the field values are bioinfo.pipeline.Input objects. These objects describe the input port behaviors.
The input port names are the expected field names of the input structure that you pass to the
block run method.
Data Types: struct
Name-value arguments for the block Function, specified as a
string scalar, character vector, string vector, or cell array of character vectors. If
you provide multiple name-value arguments, the UseFunction block sorts
and stores them alphabetically.
Data Types: char | string | cell
Names of the output arguments of the block Function, returned
as a string scalar, character vector, string vector, or cell array of character vectors.
The order of these names determines the order of outputs returned by the block.
The specified names are used as the names of required output ports of the block.
Changing this property for an existing UserFunction block renames the
block output ports and resets the order and number of output ports to match the new
value.
Data Types: char | string | cell
This property is read-only.
Output ports of the block, specified as a structure. The field
names of the structure are the names of the block output ports, and the field values are bioinfo.pipeline.Output objects. These objects describe the output port behaviors.
The field names of the output structure returned by the block run method
are the same as the output port names.
Data Types: struct
Names of the required positional input arguments to the block
Function, specified as a string scalar, character vector, string
vector, or cell array of character vectors. The order of these names determines the
order that the inputs are passed to the block Function when you
call the block run method.
The specified names are used as the names of required input ports of the block.
Changing this property for an existing UserFunction block renames the
block input ports and resets the order and number of input ports to match the new
value.
Data Types: char | string | cell
Block Function signature, specified as a string scalar or
character vector. The Signature property defines how the underlying
custom function is called when you run the block. In other words, the signature is
typically similar to what you would enter at the MATLAB® command line to run such a function. For example, the
Signature to run the aa2int function with one input and one output argument would be:
"numbers = aa2int(Seq)", where numbers is an
output variable and Seq is an input variable. For examples, see Create UserFunction Blocks For MATLAB Functions.
If you specify the Signature property, other related
properties, namely, Function,
RequiredArguments, NameValueArguments, and
OutputArguments, are automatically derived and set. Ensure that
the signature you specify is a valid MATLAB expression containing one function call.
Data Types: char | string
Object Functions
compile | Perform block-specific additional checks and validations |
copy | Copy array of handle objects |
emptyInputs | Create input structure for use with run method |
eval | Evaluate block object |
run | Run block object |
Examples
You can create a UserFunction block for any existing or custom MATLAB function.
Create UserFunction for size Function
Create a UserFunction block for the size function with a single input and output.
ufSize = bioinfo.pipeline.block.UserFunction; ufSize.Function = "size"; ufSize.RequiredArguments = "A"; ufSize.OutputArguments = "sz"
ufSize =
UserFunction with properties:
Signature: "sz = size(A)"
RequiredArguments: "A"
NameValueArguments: [0×0 string]
OutputArguments: "sz"
Function: @size
Inputs: [1×1 struct]
Outputs: [1×1 struct]
ErrorHandler: []
The UserFunction block is created. Next, create an input structure with the field name matching the input port name "A".
inStruct = struct("A",ones(2,3));Run the block using the input structure. The block result is returned as a structure with the field named "sz", which matches the output port on the block.
sizeResult = run(ufSize,inStruct)
sizeResult = struct with fields:
sz: [2 3]
Create UserFunction for align2cigar Function
Create a UserFunction block for the align2cigar function with two inputs and two outputs.
ufalign2cigar = bioinfo.pipeline.block.UserFunction; ufalign2cigar.Function = "align2cigar"; ufalign2cigar.RequiredArguments = ["alignment","ref"]; ufalign2cigar.OutputArguments = ["cigars","starts"]
ufalign2cigar =
UserFunction with properties:
Signature: "[cigars, starts] = align2cigar(alignment, ref)"
RequiredArguments: [2×1 string]
NameValueArguments: [0×1 string]
OutputArguments: [2×1 string]
Function: @align2cigar
Inputs: [1×1 struct]
Outputs: [1×1 struct]
ErrorHandler: []
The UserFunction block is created with two input ports and two output ports, which are named after the inputs (alignment and ref) and outputs (cigars and starts) that you specified.
ufalign2cigar.RequiredArguments
ans = 2×1 string
"alignment"
"ref"
ufalign2cigar.OutputArguments
ans = 2×1 string
"cigars"
"starts"
Use emptyInputs to create an input structure with the fields automatically named after the block input ports.
inStruct = emptyInputs(ufalign2cigar)
inStruct = struct with fields:
alignment: []
ref: []
Set the values of the structure fields.
inStruct.alignment = ['ACG-ATGC'; 'ACGT-TGC'; ' GTAT-C']; inStruct.ref = 'ACGTATGC';
Run the block with the input structure. The block results are returned as a structure with the fields cigars and starts.
a2cResults = run(ufalign2cigar,inStruct)
a2cResults = struct with fields:
cigars: {'3=1D4=' '4=1D3=' '4=1D1='}
starts: [1 1 3]
Create UserFunction for samread
Create a UserFunction block for the samread function that takes in multiple name-value arguments.
ufsamread = bioinfo.pipeline.block.UserFunction; ufsamread.Function = "samread"; ufsamread.RequiredArguments = "File"; ufsamread.OutputArguments = ["samData","headerData"]; ufsamread.NameValueArguments = ["blockread","tags"]
ufsamread =
UserFunction with properties:
Signature: "[samData, headerData] = samread(File, 'blockread', blockreadValue, 'tags', tagsValue)"
RequiredArguments: "File"
NameValueArguments: [2×1 string]
OutputArguments: [2×1 string]
Function: @samread
Inputs: [1×1 struct]
Outputs: [1×1 struct]
ErrorHandler: []
Use emptyInputs with IncludeOptional=true so that the structure has the fields for the required input (File) and optional name-value arguments (blockread and tags).
inStruct = emptyInputs(ufsamread,IncludeOptional=true)
inStruct = struct with fields:
File: []
blockread: []
tags: []
Set the input values. For the File input, use the provided SAM file. Read a block of sequence entries from 5 to 10 and exclude the tags.
inStruct.File = which("ex1.sam");
inStruct.blockread = [5 10];
inStruct.tags = false;Run the block. The results are returned as a structure. samData field contains sequence alignment and mapping information from the SAM file. headerData contains the header information about the SAM file.
results = run(ufsamread,inStruct)
results = struct with fields:
samData: [6×1 struct]
headerData: [1×1 struct]
results.samData(1)
ans = struct with fields:
QueryName: 'EAS56_59:8:38:671:758'
Flag: 137
ReferenceName: 'seq1'
Position: 9
MappingQuality: 99
CigarString: '35M'
MateReferenceName: '*'
MatePosition: 0
InsertSize: 0
Sequence: 'GCTCATTGTAAATGTGTGGTTTAACTCGTCCATGG'
Quality: '<<<<<<<<<<<<<<<;<;7<<<<<<<<7<<;:<5%'
results.headerData.SequenceDictionary
ans = struct with fields:
SequenceName: 'seq1'
GenomeAssemblyID: 'HG18'
SequenceLength: 62435964
Import the Pipeline and block objects needed for the example.
import bioinfo.pipeline.Pipeline import bioinfo.pipeline.block.*
Create a pipeline.
qcpipeline = Pipeline;
Select an input FASTQ file using a FileChooser block.
fastqfile = FileChooser(which("SRR005164_1_50.fastq"));Create a SeqFilter block.
sequencefilter = SeqFilter;
Define the filtering threshold value. Specifically, filter out sequences with a total of more than 10 low-quality bases, where a base is considered a low-quality base if its quality score is less than 20.
sequencefilter.Options.Threshold = [10 20];
Add the blocks to the pipeline.
addBlock(qcpipeline,[fastqfile,sequencefilter]);
Connect the output of the first block to the input of the second block. To do so, you need to first check the input and output port names of the corresponding blocks.
View the Outputs (port of the first block) and Inputs (port of the second block).
fastqfile.Outputs
ans = struct with fields:
Files: [1×1 bioinfo.pipeline.Output]
sequencefilter.Inputs
ans = struct with fields:
FASTQFiles: [1×1 bioinfo.pipeline.Input]
Connect the Files output port of the fastqfile block to the FASTQFiles port of sequencefilter block.
connect(qcpipeline,fastqfile,sequencefilter,["Files","FASTQFiles"]);
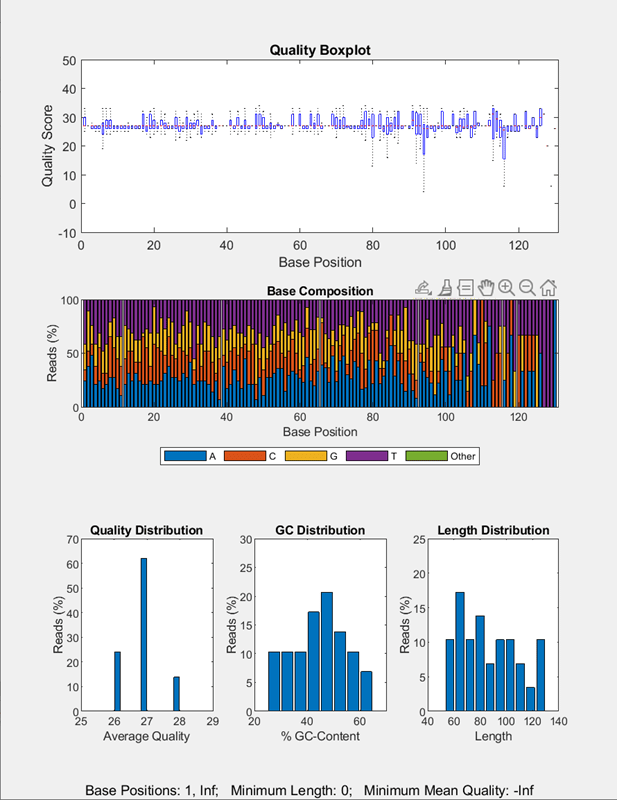
Next, create a UserFunction block that calls the seqqcplot function to plot the quality data of the filtered sequence data. In this case, inputFile is the required argument for the seqqcplot function. The required argument name can be anything as long as it is a valid variable name.
qcplot = UserFunction("seqqcplot",RequiredArguments="inputFile",OutputArguments="figureHandle");
Alternatively, you can also use dot notation to set up your UserFunction block.
qcplot = UserFunction; qcplot.RequiredArguments = "inputFile"; qcplot.Function = "seqqcplot"; qcplot.OutputArguments = "figureHandle";
Add the block.
addBlock(qcpipeline,qcplot);
Check the port names of sequencefilter block and qcplot block.
sequencefilter.Outputs
ans = struct with fields:
FilteredFASTQFiles: [1×1 bioinfo.pipeline.Output]
NumFilteredIn: [1×1 bioinfo.pipeline.Output]
NumFilteredOut: [1×1 bioinfo.pipeline.Output]
qcplot.Inputs
ans = struct with fields:
inputFile: [1×1 bioinfo.pipeline.Input]
Connect the FilteredFASTQFiles port of the sequencefilter block to the inputFile port of the qcplot block.
connect(qcpipeline,sequencefilter,qcplot,["FilteredFASTQFiles","inputFile"]);
Run the pipeline to plot the sequence quality data.
run(qcpipeline);
Version History
Introduced in R2023a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)