For most pipelines, using a combination of built-in blocks and UserFunction blocks is sufficient and recommended. However, if a block requires more complexity, such as additional configuration options or methods, you can create a custom block by subclassing the bioinfo.pipeline.Block object to implement additional capabilities.
Subclasses of the Block object must:
For instance, the following lines of code defines a custom block object named Seqlinkage which is defined as a subclass of bioinfo.pipeline.Block. For illustration purposes, the Seqlinkage block uses the seqlinkage function, which constructs a phylogenetic tree from pairwise distances as the underlying function. The function accepts a matrix or vector of pairwise distances and a distance method to use.
classdef Seqlinkage < bioinfo.pipeline.Block
% Define the block properties
properties
% Define the Method property that accepts two distance methods.
% For details on these methods, enter the following command at
% the command line: doc seqlinkage
Method {mustBeMember(Method, ["average", "weighted"])} = "average";
end
methods
% Define inputs and outputs.
function block = Seqlinkage()
import bioinfo.pipeline.Input
import bioinfo.pipeline.Output
% Define the Inputs and Outputs property of the object.
% Name the first input port asd Distances and as a required
% input that takes in a matrix or vector of pairwise distances.
block.Inputs.Distances = Input('Required',true);
% Name the second input port as Names and as an optional
% input that takes in a list of names for nodes.
block.Inputs.Names = Input('Required',false);
% Name the output port Phytree.
block.Outputs.Phytree = Output;
end
% Define custom evaluation method for the block.
function outStruct = eval(obj, inStruct)
% If the optional input (a list of node names) is passed in
if isfield(inStruct,'Names')
% Call the seqlinkage function with three inputs and save
% the returned phytree object as output.
outStruct.Phytree = seqlinkage(inStruct.Distances,obj.Method,inStruct.Names);
else
% Call seqlinkage with two inputs.
outStruct.Phytree = seqlinkage(inStruct.Distances,obj.Method);
end
end
end
end
You can save such a class definition to a separate MATLAB® program file. For this example, the above Seqlinkage class definition has already been saved and provided as Seqlinkage.m. Note that the definition file must be in the current working folder or MATLAB search path before you can use the block object in your pipeline.
Next, you can define a pipeline that uses the Seqlinkage block as follows to build a phylogenetic tree from pairwise distances.
Create three blocks needed for the workflow.
Set the input port of fastaReadBlock to a FASTA file containing amino acid sequences.
Add the blocks to the pipeline.
Connect the first two blocks.
Connect seqpdistBlock to seqlinkageBlock. Specifically, connect the output port "pairwiseDistances" of seqpdistBlock to one of the input ports of the seqlinkageBlock "Distances", as defined in the Seqlinkage.m. Note that Distances is a required input port that must have its value set. One of the valid ways is to connect to another port. For other ways to set input ports, see Satisfy Input Ports.
Connect the output port "sequences" of the fastaread block to the "Names" optional input port of seqlinkageBlock to label the leave nodes of the output phylogenetic tree.
Optionally, you can also pass in a list of node names as the value of the optional input port "Names" by setting the property seqlinkageBlock.Inputs.Names.Value.
Before you run the pipeline, ensure that the folder of the class definition file is on the MATLAB search path. The reason is because each pipeline block runs in its own folder, and the external class definition files or function files will not be detected by the pipeline unless they are on the path.
Run the pipeline.
Get the result from seqlinkageBlock, which contains a phylogenetic tree object.
linkageResult = struct with fields:
Phytree: [1×1 phytree]
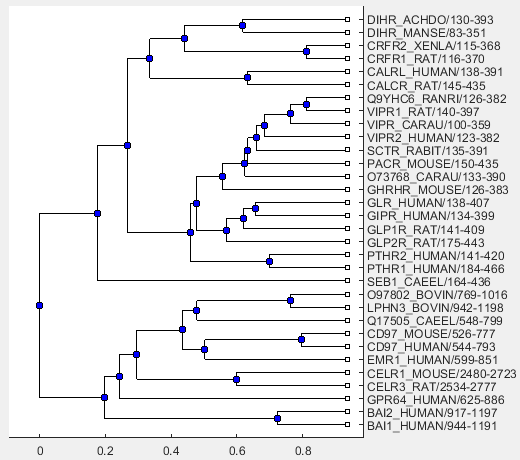
Use the following command to visualize the phylogenetic tree.
view(linkageResult.Phytree)