plot
Plot confidence interval results for model predictions
Description
fh = plot(predCI)predCI, a PredictionConfidenceInterval object or vector of objects.
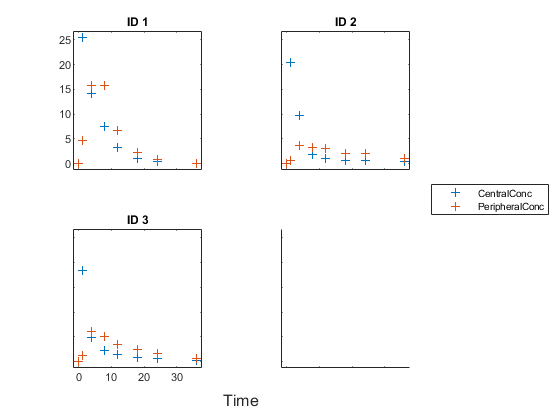
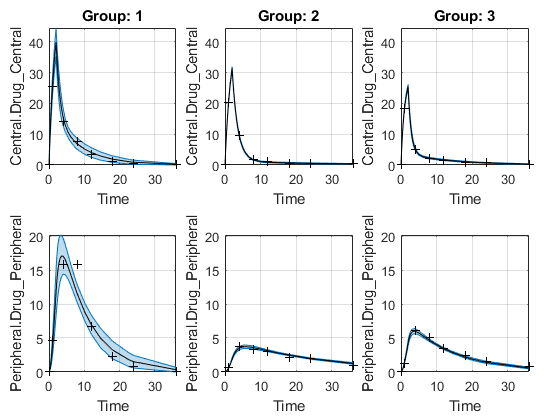
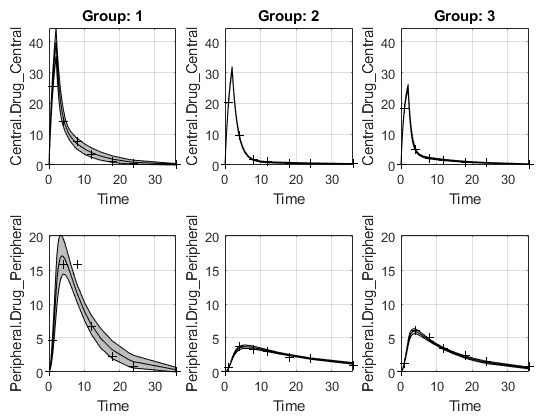
The function plots the observation data points as black plus signs and the model predictions as solid lines.
If the status of confidence interval (
predCI.Status) isconstrainedornot estimable, the function uses the second default color (red) to plot the confidence intervals.Otherwise, the function uses the first default color (blue) and plots the confidence intervals as shaded areas.
fh = plot(predCI,Color=colorValue)
Examples
Input Arguments
Output Arguments
Version History
Introduced in R2017b