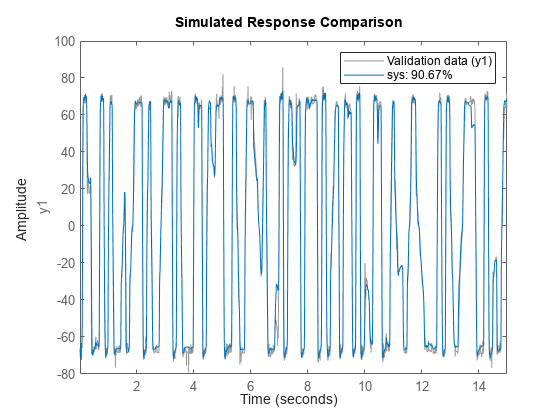
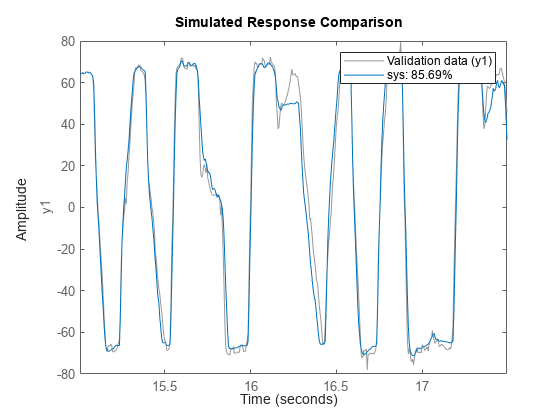
idTreeEnsemble
Decision tree ensemble mapping function for nonlinear ARX models (requires Statistics and Machine Learning Toolbox)
Since R2021b
Description
An idTreeEnsemble object implements a decision tree ensemble
model, and is a nonlinear mapping function for estimating nonlinear ARX models. This mapping
object incorporates regression tree ensembles that the mapping function creates using
Statistics and Machine Learning Toolbox™. Unlike most other mapping objects for idnlarx models, which typically contain offset, linear, and nonlinear components,
the idTreeEnsemble model contains only a nonlinear component.
![]()
Mathematically, the idTreeEnsemble object maps m
inputs x(t) =
[x1(t),x2(t),…,xm(t)]T
to a scalar output y(t) using a decision tree regression
ensemble model.
Here:
x(t) is an m-by-1 vector of inputs, or regressors.
y(t) is the scalar output.
For more information about creating regression tree ensembles, see fitrensemble (Statistics and Machine Learning Toolbox).
Use idTreeEnsemble as the value of the OutputFcn
property of an idnlarx model. For example, specify
idTreeEnsemble when you estimate an idnlarx model with the
following
command.
sys = nlarx(data,regressors,idTreeEnsemble)
nlarx estimates the model, it essentially estimates the parameters of the
idTreeEnsemble object.You can configure the idTreeEnsemble function to set options and fix
parameters. To modify the estimation options, set the option property in
E.EstimationOptions, where E is the
idTreeEnsemble object. For example, to change the fit method to
'lsboost-resampled', use E.EstimationOptions.FitMethod =
'lsboost-resampled'. To fix the values of an existing estimated
idTreeEnsemble during subsequent nlarx estimations,
set the Free property to false. To apply parallel
processing, set E.EstimationOptions.UseParallel to true.
Use evaluate to compute the output of the function for a given vector of regressor
inputs.
Creation
Description
E = idTreeEnsembleidTreeEnsemble object E with the default
estimation fit method of 'bag'. The number of regressor inputs is
determined during model estimation and the number of idTreeEnsemble
outputs is 1.
E = idTreeEnsemble(fitmethod)fitmethod.
Input Arguments
Properties
Examples
Extended Capabilities
Version History
Introduced in R2021bSee Also
nlarx | idnlarx | fitrensemble (Statistics and Machine Learning Toolbox) | evaluate
Topics
- Framework for Ensemble Learning (Statistics and Machine Learning Toolbox)
- Available Mapping Functions for Nonlinear ARX Models