Train Network with Numeric Features
This example shows how to create and train a simple neural network for deep learning feature data classification.
If you have a data set of numeric features (for example a collection of numeric data without spatial or time dimensions), then you can train a deep learning network using a feature input layer. For an example showing how to train a network for image classification, see Create Simple Deep Learning Neural Network for Classification.
This example shows how to train a network to classify the gear tooth condition of a transmission system given a mixture of numeric sensor readings, statistics, and categorical labels.
Load Data
Load the transmission casing dataset for training. The data set consists of 208 synthetic readings of a transmission system consisting of 18 numeric readings and three categorical labels:
SigMean— Vibration signal meanSigMedian— Vibration signal medianSigRMS— Vibration signal RMSSigVar— Vibration signal varianceSigPeak— Vibration signal peakSigPeak2Peak— Vibration signal peak to peakSigSkewness— Vibration signal skewnessSigKurtosis— Vibration signal kurtosisSigCrestFactor— Vibration signal crest factorSigMAD— Vibration signal MADSigRangeCumSum— Vibration signal range cumulative sumSigCorrDimension— Vibration signal correlation dimensionSigApproxEntropy— Vibration signal approximate entropySigLyapExponent— Vibration signal Lyap exponentPeakFreq— Peak frequency.HighFreqPower— High frequency powerEnvPower— Environment powerPeakSpecKurtosis— Peak frequency of spectral kurtosisSensorCondition— Condition of sensor, specified as "Sensor Drift" or "No Sensor Drift"ShaftCondition— Condition of shaft, specified as "Shaft Wear" or "No Shaft Wear"GearToothCondition— Condition of gear teeth, specified as "Tooth Fault" or "No Tooth Fault"
Read the transmission casing data from the CSV file "transmissionCasingData.csv".
filename = "transmissionCasingData.csv"; tbl = readtable(filename,TextType="String");
Convert the labels for prediction to categorical using the convertvars function.
labelName = "GearToothCondition"; tbl = convertvars(tbl,labelName,"categorical");
View the first few rows of the table.
head(tbl)
SigMean SigMedian SigRMS SigVar SigPeak SigPeak2Peak SigSkewness SigKurtosis SigCrestFactor SigMAD SigRangeCumSum SigCorrDimension SigApproxEntropy SigLyapExponent PeakFreq HighFreqPower EnvPower PeakSpecKurtosis SensorCondition ShaftCondition GearToothCondition
________ _________ ______ _______ _______ ____________ ___________ ___________ ______________ _______ ______________ ________________ ________________ _______________ ________ _____________ ________ ________________ _______________ _______________ __________________
-0.94876 -0.9722 1.3726 0.98387 0.81571 3.6314 -0.041525 2.2666 2.0514 0.8081 28562 1.1429 0.031581 79.931 0 6.75e-06 3.23e-07 162.13 "Sensor Drift" "No Shaft Wear" No Tooth Fault
-0.97537 -0.98958 1.3937 0.99105 0.81571 3.6314 -0.023777 2.2598 2.0203 0.81017 29418 1.1362 0.037835 70.325 0 5.08e-08 9.16e-08 226.12 "Sensor Drift" "No Shaft Wear" No Tooth Fault
1.0502 1.0267 1.4449 0.98491 2.8157 3.6314 -0.04162 2.2658 1.9487 0.80853 31710 1.1479 0.031565 125.19 0 6.74e-06 2.85e-07 162.13 "Sensor Drift" "Shaft Wear" No Tooth Fault
1.0227 1.0045 1.4288 0.99553 2.8157 3.6314 -0.016356 2.2483 1.9707 0.81324 30984 1.1472 0.032088 112.5 0 4.99e-06 2.4e-07 162.13 "Sensor Drift" "Shaft Wear" No Tooth Fault
1.0123 1.0024 1.4202 0.99233 2.8157 3.6314 -0.014701 2.2542 1.9826 0.81156 30661 1.1469 0.03287 108.86 0 3.62e-06 2.28e-07 230.39 "Sensor Drift" "Shaft Wear" No Tooth Fault
1.0275 1.0102 1.4338 1.0001 2.8157 3.6314 -0.02659 2.2439 1.9638 0.81589 31102 1.0985 0.033427 64.576 0 2.55e-06 1.65e-07 230.39 "Sensor Drift" "Shaft Wear" No Tooth Fault
1.0464 1.0275 1.4477 1.0011 2.8157 3.6314 -0.042849 2.2455 1.9449 0.81595 31665 1.1417 0.034159 98.838 0 1.73e-06 1.55e-07 230.39 "Sensor Drift" "Shaft Wear" No Tooth Fault
1.0459 1.0257 1.4402 0.98047 2.8157 3.6314 -0.035405 2.2757 1.955 0.80583 31554 1.1345 0.0353 44.223 0 1.11e-06 1.39e-07 230.39 "Sensor Drift" "Shaft Wear" No Tooth Fault
To train a network using categorical features, you must first convert the categorical features to numeric. First, convert the categorical predictors to categorical using the convertvars function by specifying a string array containing the names of all the categorical input variables. In this data set, there are two categorical features with names "SensorCondition" and "ShaftCondition".
categoricalInputNames = ["SensorCondition" "ShaftCondition"]; tbl = convertvars(tbl,categoricalInputNames,"categorical");
Loop over the categorical input variables. For each variable:
Convert the categorical values to one-hot encoded vectors using the
onehotencodefunction.Add the one-hot vectors to the table using the
addvarsfunction. Specify to insert the vectors after the column containing the corresponding categorical data.Remove the corresponding column containing the categorical data.
for i = 1:numel(categoricalInputNames) name = categoricalInputNames(i); oh = onehotencode(tbl(:,name)); tbl = addvars(tbl,oh,After=name); tbl(:,name) = []; end
Split the vectors into separate columns using the splitvars function.
tbl = splitvars(tbl);
View the first few rows of the table. Notice that the categorical predictors have been split into multiple columns with the categorical values as the variable names.
head(tbl)
SigMean SigMedian SigRMS SigVar SigPeak SigPeak2Peak SigSkewness SigKurtosis SigCrestFactor SigMAD SigRangeCumSum SigCorrDimension SigApproxEntropy SigLyapExponent PeakFreq HighFreqPower EnvPower PeakSpecKurtosis No Sensor Drift Sensor Drift No Shaft Wear Shaft Wear GearToothCondition
________ _________ ______ _______ _______ ____________ ___________ ___________ ______________ _______ ______________ ________________ ________________ _______________ ________ _____________ ________ ________________ _______________ ____________ _____________ __________ __________________
-0.94876 -0.9722 1.3726 0.98387 0.81571 3.6314 -0.041525 2.2666 2.0514 0.8081 28562 1.1429 0.031581 79.931 0 6.75e-06 3.23e-07 162.13 0 1 1 0 No Tooth Fault
-0.97537 -0.98958 1.3937 0.99105 0.81571 3.6314 -0.023777 2.2598 2.0203 0.81017 29418 1.1362 0.037835 70.325 0 5.08e-08 9.16e-08 226.12 0 1 1 0 No Tooth Fault
1.0502 1.0267 1.4449 0.98491 2.8157 3.6314 -0.04162 2.2658 1.9487 0.80853 31710 1.1479 0.031565 125.19 0 6.74e-06 2.85e-07 162.13 0 1 0 1 No Tooth Fault
1.0227 1.0045 1.4288 0.99553 2.8157 3.6314 -0.016356 2.2483 1.9707 0.81324 30984 1.1472 0.032088 112.5 0 4.99e-06 2.4e-07 162.13 0 1 0 1 No Tooth Fault
1.0123 1.0024 1.4202 0.99233 2.8157 3.6314 -0.014701 2.2542 1.9826 0.81156 30661 1.1469 0.03287 108.86 0 3.62e-06 2.28e-07 230.39 0 1 0 1 No Tooth Fault
1.0275 1.0102 1.4338 1.0001 2.8157 3.6314 -0.02659 2.2439 1.9638 0.81589 31102 1.0985 0.033427 64.576 0 2.55e-06 1.65e-07 230.39 0 1 0 1 No Tooth Fault
1.0464 1.0275 1.4477 1.0011 2.8157 3.6314 -0.042849 2.2455 1.9449 0.81595 31665 1.1417 0.034159 98.838 0 1.73e-06 1.55e-07 230.39 0 1 0 1 No Tooth Fault
1.0459 1.0257 1.4402 0.98047 2.8157 3.6314 -0.035405 2.2757 1.955 0.80583 31554 1.1345 0.0353 44.223 0 1.11e-06 1.39e-07 230.39 0 1 0 1 No Tooth Fault
View the class names of the data set.
classNames = categories(tbl{:,labelName})classNames = 2×1 cell
{'No Tooth Fault'}
{'Tooth Fault' }
Split Data Set into Training and Validation Sets
Partition the data set into training, validation, and test partitions. Set aside 15% of the data for validation, and 15% for testing.
View the number of observations in the dataset.
numObservations = size(tbl,1)
numObservations = 208
Determine the number of observations for each partition.
numObservationsTrain = floor(0.7*numObservations)
numObservationsTrain = 145
numObservationsValidation = floor(0.15*numObservations)
numObservationsValidation = 31
numObservationsTest = numObservations - numObservationsTrain - numObservationsValidation
numObservationsTest = 32
Create an array of random indices corresponding to the observations and partition it using the partition sizes.
idx = randperm(numObservations); idxTrain = idx(1:numObservationsTrain); idxValidation = idx(numObservationsTrain+1:numObservationsTrain+numObservationsValidation); idxTest = idx(numObservationsTrain+numObservationsValidation+1:end);
Partition the table of data into training, validation, and testing partitions using the indices.
tblTrain = tbl(idxTrain,:); tblValidation = tbl(idxValidation,:); tblTest = tbl(idxTest,:);
Define Network Architecture
Define the network for classification.
Define a network with a feature input layer and specify the number of features. Also, configure the input layer to normalize the data using Z-score normalization. Next, include a fully connected layer with output size 50 followed by a batch normalization layer and a ReLU layer. For classification, specify another fully connected layer with output size corresponding to the number of classes, followed by a softmax layer.
numFeatures = size(tbl,2) - 1;
numClasses = numel(classNames);
layers = [
featureInputLayer(numFeatures,Normalization="zscore")
fullyConnectedLayer(50)
batchNormalizationLayer
reluLayer
fullyConnectedLayer(numClasses)
softmaxLayer];Specify Training Options
Specify the training options.
Train the network using Adam.
Train using mini-batches of size 16.
Shuffle the data every epoch.
Monitor the network accuracy during training by specifying validation data.
Display the training progress in a plot and suppress the verbose command window output.
The software trains the network on the training data and calculates the accuracy on the validation data at regular intervals during training. The validation data is not used to update the network weights.
miniBatchSize = 16; options = trainingOptions("adam", ... MiniBatchSize=miniBatchSize, ... Shuffle="every-epoch", ... ValidationData=tblValidation, ... Plots="training-progress", ... Metrics="accuracy", ... Verbose=false);
Train Network
Train the network using the architecture defined by layers, the training data, and the training options. By default, trainnet uses a GPU if one is available, otherwise, it uses a CPU. Training on a GPU requires Parallel Computing Toolbox™ and a supported GPU device. For information on supported devices, see GPU Computing Requirements (Parallel Computing Toolbox). You can also specify the execution environment by using the ExecutionEnvironment name-value argument of trainingOptions.
The training progress plot shows the mini-batch loss and accuracy and the validation loss and accuracy. For more information on the training progress plot, see Monitor Deep Learning Training Progress.
net = trainnet(tblTrain,layers,"crossentropy",options);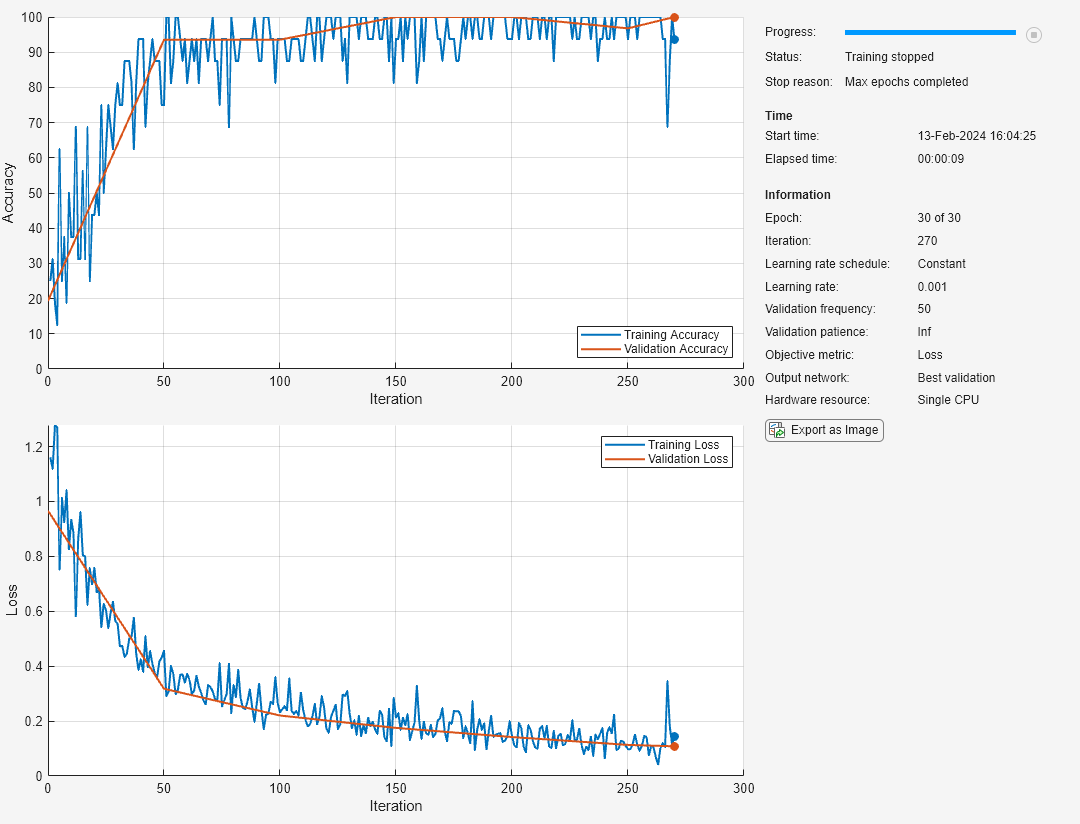
Test Network
Predict the labels of the test data using the trained network and calculate the accuracy. Specify the same mini-batch size used for training.
scores = minibatchpredict(net,tblTest(:,1:end-1),MiniBatchSize=miniBatchSize); YPred = scores2label(scores,classNames);
Calculate the classification accuracy. The accuracy is the proportion of the labels that the network predicts correctly.
YTest = tblTest{:,labelName};
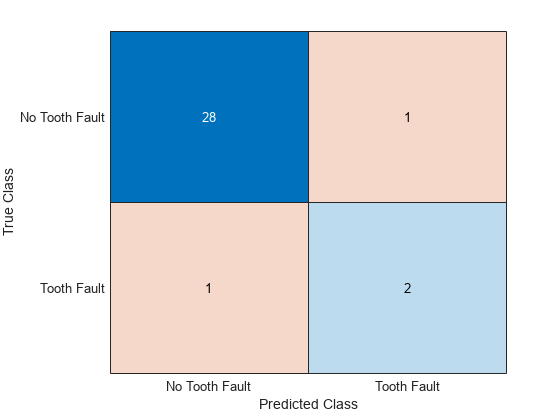
accuracy = sum(YPred == YTest)/numel(YTest)accuracy = 0.9375
View the results in a confusion matrix.
figure confusionchart(YTest,YPred)
See Also
trainnet | trainingOptions | dlnetwork | fullyConnectedLayer | Deep Network
Designer | featureInputLayer