Identify Vegetation Regions Using Interactive NDVI Thresholding
This example shows how to identify the types of vegetations regions in a hyperspectral image through interactive thresholding of a normalized difference vegetation index (NDVI) map. The NDVI map of a hyperspectral dataset indicates the density of vegetation in various regions of the hyperspectral data. The NDVI value is computed using the near-infrared (NIR) and visible red (R) spectral band images from the hyperspectral data cube.
The NDVI value of a pixel is a scalar from -1 to 1. The pixels in regions with healthy or dense vegetation reflect more NIR light, resulting in high NDVI values. The pixels in regions with unhealthy vegetation or barren land absorb more NIR light, resulting in low or negative NDVI values. Based on its NDVI value, you can identify vegetation in a region as dense vegetation, moderate vegetation, sparse vegetation, or no vegetation. These are the typical NDVI value range for each type of region:
Dense vegetation - Greater than or equal to 0.6
Moderate vegetation - In the range [0.4, 0.6)
Sparse vegetation - In the range [0.2 0.4)
No vegetation - Below 0.2
You can segment the desired vegetation regions by performing thresholding using the NDVI values. In this example, you will interactively select and change the threshold values to identify different vegetation regions in a hyperspectral data cube based on their NDVI values.
This example requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons. The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Read Hyperspectral Data
Read hyperspectral data from an ENVI format file into the workspace. This example uses a data sample from the Pavia dataset, which contains both vegetation and barren regions.
hcube = imhypercube("paviaU.dat");Compute NDVI
Compute the NDVI value for each pixel in the data cube by using the ndvi function. The function outputs a 2-D image in which the value of each pixel is the NDVI value for the corresponding pixel in the hyperspectral data cube.
ndviImg = ndvi(hcube);
Identify Vegetation Regions Using Thresholding
Identify different regions in the hyperspectral data using multilevel thresholding. Define a label matrix to assign label values to pixels based on specified threshold values. You can set the thresholds based on the computed NDVI values.
Label value
1- Specify the threshold value as 0.6, and find the pixels with NDVI values greater than or equal to the threshold. These are dense vegetation pixels.Label value
2- Specify a lower threshold limit of 0.4 and an upper threshold limit of 0.6. Find the pixels with NDVI values greater than or equal to 0.4 and less than 0.6. These are moderate vegetation pixels.Label value
3- Specify a lower threshold limit of 0.2 and an upper threshold limit of 0.4. Find the pixels with NDVI values greater than or equal to 0.2 and less than 0.4. These are the sparse vegetation pixels.Label value
4- Specify the threshold value as 0.2, and find the pixels with NDVI values less than the threshold. These are no vegetation pixels.
L = zeros(size(ndviImg)); L(ndviImg >= 0.6) = 1; L(ndviImg >= 0.4 & ndviImg < 0.6) = 2; L(ndviImg >= 0.2 & ndviImg < 0.4) = 3; L(ndviImg < 0.2) = 4;
Estimate a contrast-stretched RGB image from the original data cube.
rgbImg = colorize(hcube,Method="rgb",ContrastStretching=true);Define a colormap to display each value in the label matrix in a different color. Overlay the label matrix on the RGB image.
cmap = [0 1 0; 0 0 1; 1 1 0; 1 0 0]; overlayImg = labeloverlay(rgbImg,L,Colormap=cmap);
Create Interactive Interface for NDVI Thresholding
To build an interactive interface, first create a figure window. Then add two panels to the figure window, for displaying the input image and the overlaid image side by side.
h = uifigure(Name="Interactive NDVI Thresholding",Position=[200,50,1000,700]); viewPanel1 = uipanel(h,Position=[2 220 400 450],Title="Input Image"); ax1 = axes(viewPanel1); image(rgbImg,Parent=ax1) viewPanel2 = uipanel(h,Position=[400 220 400 450],Title="Types of Vegetation Regions in Input Image"); ax2 = axes(viewPanel2); image(overlayImg,Parent=ax2)
Annotate the figure window with the color for each label and its associated vegetation density. The colormap value for dense vegetation is green, moderate vegetation is blue, sparse vegetation is yellow, and no vegetation is red.
annotation(h,"rectangle",[0.82 0.82 0.03 0.03],Color=[0 1 0],FaceColor=[0 1 0]); annotation(h,"rectangle",[0.82 0.77 0.03 0.03],Color=[0 0 1],FaceColor=[0 0 1]); annotation(h,"rectangle",[0.82 0.72 0.03 0.03],Color=[1 1 0],FaceColor=[1 1 0]); annotation(h,"rectangle",[0.82 0.67 0.03 0.03],Color=[1 0 0],FaceColor=[1 0 0]); annotation(h,"textbox",[0.85 0.80 0.9 0.05],EdgeColor="None",String="Dense Vegetation"); annotation(h,"textbox",[0.85 0.75 0.9 0.05],EdgeColor="None",String="Moderate Vegetation"); annotation(h,"textbox",[0.85 0.70 0.9 0.05],EdgeColor="None",String="Sparse Vegetation"); annotation(h,"textbox",[0.85 0.65 0.9 0.05],EdgeColor="None",String="No Vegetation");
Create sliders for interactively changing the thresholds. Add a slider for adjusting the minimum threshold value and a slider for adjusting the maximum threshold value.
slidePanel1 = uipanel(h,Position=[400,120,400,70],Title="Minimum Threshold Value"); minsld = uislider(slidePanel1,Position=[30,40,350,3],Value=-1,Limits=[-1 1],MajorTicks=-1:0.4:1); slidePanel2 = uipanel(h,Position=[400,30,400,70],Title="Maximum Threshold Value"); maxsld = uislider(slidePanel2,Position=[30,35,350,3],Value=1,Limits=[-1 1],MajorTicks=-1:0.4:1);
Change Threshold Interactively
Use the function ndviThreshold function, defined at the end of this example, to change the minimum and maximum threshold limits. When you move the slider thumb and release the mouse button, the ValueChangedFcn callback updates the slider value and sets the slider value as the new threshold. You must call the ndviThreshold function separately for the minimum threshold slider and maximum threshold slider. Change the threshold limits by adjusting the sliders. This enables you to inspect the types of vegetation regions within your specified threshold limits.
minsld.ValueChangedFcn = @(es,ed) ndviThreshold(minsld,maxsld,ndviImg,rgbImg,ax2,cmap); maxsld.ValueChangedFcn = @(es,ed) ndviThreshold(minsld,maxsld,ndviImg,rgbImg,ax2,cmap);
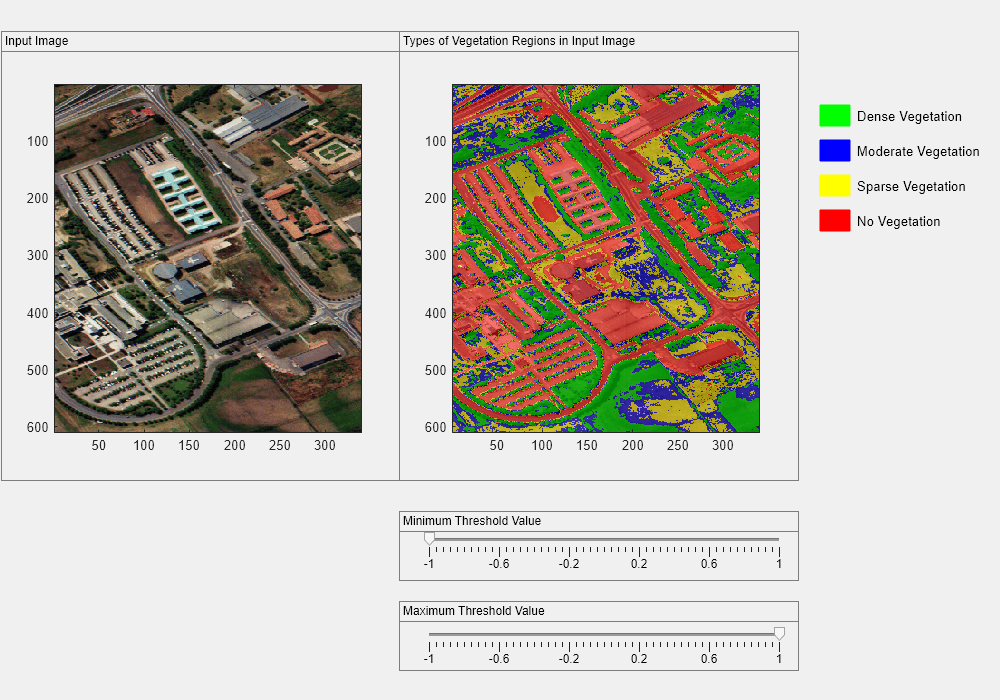
The ndviThreshold function generates a new label matrix using the updated threshold values and dynamically updates the overlaid image in the figure window.
Create Callback Function
Create callback function to interactively change the threshold limits and dynamically update the results.
function ndviThreshold(minsld,maxsld,ndviImg,rgbImg,ax2,cmap) L = zeros(size(ndviImg)); minth = round(minsld.Value,2); maxth = round(maxsld.Value,2); if minth > maxth error("Minimum threshold value must be less than the maximum threshold value") end if minth >= 0.6 % Label 1 for Dense Vegetation L(ndviImg >= minth & ndviImg <= maxth) = 1; overlayImg = labeloverlay(rgbImg,L,Colormap=cmap); elseif minth >= 0.4 && minth < 0.6 % Label 1 for Dense Vegetation % Label 2 for Moderate Vegetation if maxth >= 0.6 L(ndviImg >= minth & ndviImg < 0.6) = 2; L(ndviImg >= 0.6 & ndviImg <= maxth) = 1; else L(ndviImg >= minth & ndviImg < maxth) = 2; end overlayImg = labeloverlay(rgbImg,L,Colormap=cmap); elseif minth >= 0.2 && minth <0.4 % Label 1 for Dense Vegetation % Label 2 for Moderate Vegetation % Label 3 for Sparse vegetation if maxth < 0.4 L(ndviImg >= minth & ndviImg <= maxth) = 3; elseif maxth >=0.4 && maxth < 0.6 L(ndviImg >= minth & ndviImg < 0.4) = 3; L(ndviImg >= 0.4 & ndviImg <= maxth) = 2; elseif maxth >= 0.6 L(ndviImg >= minth & ndviImg < 0.4) = 3; L(ndviImg >= 0.4 & ndviImg < 0.6) = 2; L(ndviImg >= 0.6 & ndviImg <= maxth) = 1; end overlayImg = labeloverlay(rgbImg,L,Colormap=cmap); elseif minth < 0.2 % Label 1 for Dense Vegetation % Label 2 for Moderate Vegetation % Label 3 for Sparse vegetation % Label 4 for No Vegetation L(ndviImg >= minth & ndviImg < 0.2) = 4; if maxth >= 0.6 L(ndviImg >= 0.6 & ndviImg <= maxth) = 1; L(ndviImg >= 0.4 & ndviImg < 0.6) = 2; L(ndviImg >= 0.2 & ndviImg < 0.4) = 3; elseif maxth >=0.4 && maxth < 0.6 L(ndviImg >= 0.4 & ndviImg <= maxth) = 2; L(ndviImg >= 0.2 & ndviImg < 0.4) = 3; elseif maxth >=0.2 && maxth < 0.4 L(ndviImg >= maxth & ndviImg < 0.4) = 3; end overlayImg = labeloverlay(rgbImg,L,Colormap=cmap); elseif minth == maxth if maxth < 0.2 L(ndviImg == minth) = 4; elseif maxth >=0.4 && maxth < 0.6 L(ndviImg == maxth) = 2; elseif maxth >=0.2 && maxth < 0.4 L(ndviImg == maxth) = 3; elseif maxth >= 0.6 L(ndviImg == maxth) = 1; end overlayImg = labeloverlay(rgbImg,L,Colormap=cmap); end % Display the overlay image. image(overlayImg,Parent=ax2); % Store updated labeled image minsld.UserData = L; maxsld.UserData = L; end
References
[1] J.W. Rouse, R.H. Hass, J.A. Schell, and D.W. Deering. "Monitoring Vegetation Systems in the Great Plains with ERTS." In Proceedings of the Third Earth Resources Technology Satellite- 1 Symposium, 1:309–17. Greenbelt, NASA SP-351, Washington, DC, 1973.
[2] Haboudane, D. "Hyperspectral Vegetation Indices and Novel Algorithms for Predicting Green LAI of Crop Canopies: Modeling and Validation in the Context of Precision Agriculture." Remote Sensing of Environment 90, no. 3 (April 15, 2004): 337–52. https://doi.org/10.1016/j.rse.2003.12.013.
[3] "Change Detection of Vegetation Cover by NDVI Technique on Catchment Area of the Panchet Hill Dam, India." International Journal of Research in Geography 2, no. 3 (2016). https://doi.org/10.20431/2454-8685.0203002.
See Also
hypercube | colorize | uifigure | uipanel | uislider | labeloverlay