Representing All Data in an ExpressionSet Object
Overview of ExpressionSet Objects
You can store all microarray experiment data and information in one object by assembling the following into an ExpressionSet object:
One ExptData object containing expression values from a microarray experiment in one or more DataMatrix objects
One MetaData object containing sample metadata in two dataset arrays
One MetaData object containing feature metadata in two dataset arrays
One MIAME object containing experiment descriptions
The following graphic illustrates a typical ExpressionSet object and its component objects.
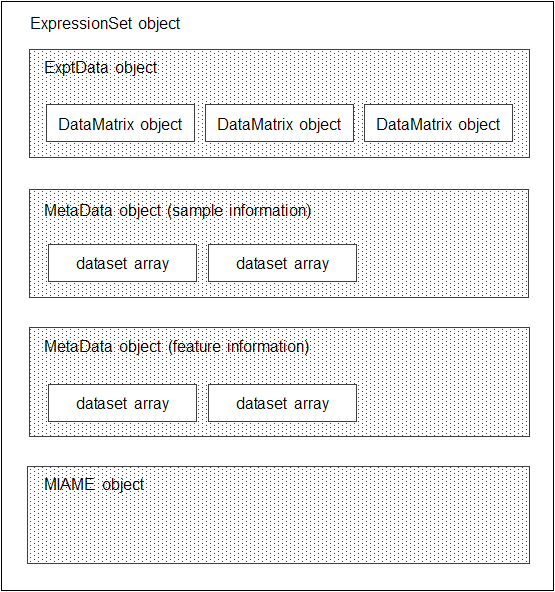
Each element (DataMatrix object) in the ExpressionSet object
has an element name. Also, there is always one DataMatrix object whose
element name is Expressions.
An ExpressionSet object lets you store, manage, and subset the data from a microarray gene expression experiment. An ExpressionSet object includes properties and methods that let you access, retrieve, and change data, metadata, and other information about the microarray experiment. These properties and methods are useful to view and analyze the data. For a list of the properties and methods, see ExpressionSet class.
Constructing ExpressionSet Objects
Note
The following procedure assumes you have executed the example code in the previous sections:
Import the
biomapackage so that theExpressionSetconstructor function is available.import bioma.*
Construct an ExpressionSet object from
EDObj, an ExptData object,MDObj2, a MetaData object containing sample variable information, andMIAMEObj, a MIAME object.ESObj = ExpressionSet(EDObj, 'SData', MDObj2, 'EInfo', MIAMEObj1);
Display information about the ExpressionSet object,
ESObj.ESObj ExpressionSet Experiment Data: 500 features, 26 samples Element names: Expressions Sample Data: Sample names: A, B, ...,Z (26 total) Sample variable names and meta information: Gender: Gender of the mouse in study Age: The number of weeks since mouse birth Type: Genetic characters Strain: The mouse strain Source: The tissue source for RNA collection Feature Data: none Experiment Information: use 'exptInfo(obj)'
For complete information on constructing ExpressionSet objects, see ExpressionSet class.
Using Properties of an ExpressionSet Object
To access properties of an ExpressionSet object, use the following syntax:
objectname.propertyname
For example, to determine the number of samples in an ExpressionSet object:
ESObj.NSamples
ans =
26
Note
Property names are case sensitive. For a list and description of all properties of an ExpressionSet object, see ExpressionSet class.
Using Methods of an ExpressionSet Object
To use methods of an ExpressionSet object, use either of the following syntaxes:
objectname.methodname
or
methodname(objectname)
For example, to retrieve the sample variable names from an ExpressionSet object:
ESObj.sampleVarNames
ans =
'Gender' 'Age' 'Type' 'Strain' 'Source'To retrieve the experiment information contained in an ExpressionSet object:
exptInfo(ESObj)
ans =
Experiment description
Author name: Mika,,Silvennoinen
Riikka,,Kivelä
Maarit,,Lehti
Anna-Maria,,Touvras
Jyrki,,Komulainen
Veikko,,Vihko
Heikki,,Kainulainen
Laboratory: XYZ Lab
Contact information: Mika,,Silvennoinen
URL:
PubMedIDs: 17003243
Abstract: A 90 word abstract is available Use the Abstract property.
Experiment Design: A 234 word summary is available Use the ExptDesign property.
Other notes:
[1x80 char]Note
For a complete list of methods of an ExpressionSet object, see ExpressionSet class.
